Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
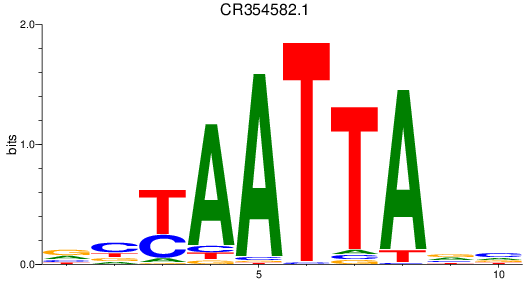
Results for CR354582.1
Z-value: 0.98
Transcription factors associated with CR354582.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CR354582.1
|
ENSDARG00000110598 | ENSDARG00000110598 |
Activity profile of CR354582.1 motif
Sorted Z-values of CR354582.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_11385155 | 5.51 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr11_+_30244356 | 5.15 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr3_-_50443607 | 4.98 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr3_-_31079186 | 4.75 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr19_+_10339538 | 3.49 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr20_-_40755614 | 3.13 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr22_+_28337204 | 3.10 |
ENSDART00000163352
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr25_-_10503043 | 3.09 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr2_+_39021282 | 3.09 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr22_+_28337429 | 3.07 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr18_+_1837668 | 2.91 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr19_+_9174166 | 2.77 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr5_-_41494831 | 2.76 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr5_-_50992690 | 2.74 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr11_+_38280454 | 2.49 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr23_+_40460333 | 2.40 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr5_+_64732270 | 2.16 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr13_+_29462249 | 2.12 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr23_-_19230627 | 2.05 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr25_-_25384045 | 2.03 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr6_+_9870192 | 2.03 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr2_+_50608099 | 2.02 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr4_+_11384891 | 2.02 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr10_+_25726694 | 2.01 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr14_-_2933185 | 2.01 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr21_-_42100471 | 2.01 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr4_-_4387012 | 2.01 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr19_-_13808630 | 1.98 |
ENSDART00000166895
ENSDART00000187670 |
ctgfb
|
connective tissue growth factor b |
| chr1_-_50859053 | 1.98 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr19_+_43297546 | 1.92 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr8_-_23780334 | 1.89 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr5_+_64732036 | 1.80 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr16_+_5774977 | 1.78 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr8_-_14126646 | 1.74 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr6_-_55585423 | 1.70 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr4_+_21129752 | 1.70 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr14_+_40874608 | 1.70 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr17_-_200316 | 1.68 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr21_-_18993110 | 1.63 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr4_+_12612723 | 1.63 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr21_-_22827548 | 1.56 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr23_+_33963619 | 1.55 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr21_-_27185915 | 1.54 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr15_+_47903864 | 1.49 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr11_-_44876005 | 1.47 |
ENSDART00000192006
|
opn6a
|
opsin 6, group member a |
| chr13_-_44630111 | 1.46 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr19_-_657439 | 1.44 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr1_+_55755304 | 1.38 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr23_+_44741500 | 1.35 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr14_+_25817628 | 1.35 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr11_+_7264457 | 1.34 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr24_-_24282427 | 1.33 |
ENSDART00000123299
|
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr12_+_16168342 | 1.32 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr9_-_22057658 | 1.30 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr22_-_21897203 | 1.28 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr17_-_23616626 | 1.26 |
ENSDART00000104730
|
ifit14
|
interferon-induced protein with tetratricopeptide repeats 14 |
| chr23_+_26946744 | 1.24 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr16_+_31804590 | 1.23 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr22_+_17205608 | 1.22 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr3_+_33341640 | 1.18 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr20_-_46362606 | 1.17 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr24_-_39518599 | 1.11 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr20_-_9462433 | 1.09 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr13_+_38814521 | 1.07 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr3_+_34919810 | 1.06 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr11_-_42554290 | 1.04 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr22_-_16416882 | 1.02 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr16_+_17389116 | 1.01 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr12_-_19007834 | 1.01 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr7_+_30787903 | 1.00 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr10_+_21867307 | 1.00 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr21_+_11244068 | 0.99 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr18_-_43884044 | 0.99 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr24_+_13316737 | 0.99 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr11_+_28476298 | 0.98 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr1_+_52929185 | 0.95 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr23_+_28582865 | 0.95 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr19_-_5103141 | 0.94 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr22_-_12160283 | 0.93 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr4_+_3980247 | 0.92 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr21_-_5205617 | 0.92 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr19_-_20113696 | 0.91 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr10_-_13343831 | 0.89 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr7_-_72261721 | 0.89 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr12_+_24342303 | 0.89 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr5_+_37903790 | 0.87 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr22_+_5176693 | 0.86 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr19_-_5103313 | 0.85 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr18_-_1185772 | 0.85 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr6_-_6487876 | 0.85 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr23_+_20110086 | 0.84 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr2_-_23778180 | 0.83 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr5_+_30624183 | 0.81 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr22_+_20427170 | 0.81 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr5_-_67629263 | 0.78 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr23_-_14990865 | 0.77 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr15_-_34785594 | 0.75 |
ENSDART00000154256
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr24_-_31425799 | 0.74 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr10_-_20696518 | 0.74 |
ENSDART00000064581
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr20_-_9436521 | 0.73 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr12_+_10631266 | 0.72 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr24_-_31843173 | 0.72 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr6_+_3280939 | 0.72 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr5_-_9625459 | 0.69 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr20_-_14925281 | 0.69 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr25_-_13842618 | 0.68 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr3_+_17537352 | 0.68 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr11_-_6048490 | 0.67 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr22_-_10121880 | 0.67 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr15_-_22074315 | 0.67 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr2_+_44615000 | 0.67 |
ENSDART00000188826
ENSDART00000113232 |
yeats2
|
YEATS domain containing 2 |
| chr14_+_8940326 | 0.66 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr8_+_20776654 | 0.66 |
ENSDART00000135850
|
nfic
|
nuclear factor I/C |
| chr24_+_16547035 | 0.66 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr9_+_50001746 | 0.65 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr12_+_2446837 | 0.65 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr10_-_26744131 | 0.65 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr10_+_16584382 | 0.65 |
ENSDART00000112039
|
CR790388.1
|
|
| chr12_+_20352400 | 0.64 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr20_-_16171297 | 0.64 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr10_+_42423318 | 0.63 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr22_-_5518117 | 0.62 |
ENSDART00000164613
|
CABZ01064972.1
|
|
| chr8_-_21142550 | 0.61 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr22_-_10586191 | 0.60 |
ENSDART00000148418
|
si:dkey-42i9.16
|
si:dkey-42i9.16 |
| chr21_-_14174786 | 0.60 |
ENSDART00000145366
|
whrna
|
whirlin a |
| chr1_+_18811679 | 0.59 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr1_-_5455498 | 0.59 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr3_-_23406964 | 0.58 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr2_-_15324837 | 0.57 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr10_-_15963903 | 0.55 |
ENSDART00000142357
|
si:dkey-3h23.3
|
si:dkey-3h23.3 |
| chr21_+_32820175 | 0.54 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr21_-_3853204 | 0.53 |
ENSDART00000188829
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr2_+_8779164 | 0.51 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr20_-_32270866 | 0.51 |
ENSDART00000153140
|
armc2
|
armadillo repeat containing 2 |
| chr18_-_2433011 | 0.50 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr5_-_15851953 | 0.49 |
ENSDART00000173101
|
si:dkey-1k23.3
|
si:dkey-1k23.3 |
| chr1_-_18811517 | 0.49 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr4_+_9400012 | 0.48 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr13_+_38817871 | 0.47 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr10_-_26512993 | 0.47 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr19_+_31904836 | 0.47 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr10_-_26512742 | 0.46 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr3_+_18398876 | 0.43 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr11_-_29768054 | 0.43 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr20_-_14924858 | 0.43 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr6_+_39232245 | 0.43 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr10_-_32494304 | 0.42 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr17_+_3379673 | 0.41 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr19_-_32710922 | 0.41 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr9_-_50001606 | 0.40 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr9_+_41156818 | 0.40 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr10_-_33297864 | 0.39 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr22_+_5176255 | 0.38 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr23_-_1658980 | 0.37 |
ENSDART00000186227
|
CU693481.1
|
|
| chr12_-_33357655 | 0.37 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr24_+_17270129 | 0.37 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr14_+_30795559 | 0.36 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr24_-_4782052 | 0.36 |
ENSDART00000149911
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr6_+_9289802 | 0.36 |
ENSDART00000188650
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr1_-_17715493 | 0.35 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr3_-_34084387 | 0.34 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr14_+_33329420 | 0.33 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr3_+_17933553 | 0.33 |
ENSDART00000167731
ENSDART00000165644 |
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr14_+_33329761 | 0.33 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr22_+_19407531 | 0.32 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr12_+_30367079 | 0.32 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr7_-_12464412 | 0.31 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr9_+_22485343 | 0.29 |
ENSDART00000146028
|
dgkg
|
diacylglycerol kinase, gamma |
| chr14_+_23717165 | 0.25 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr7_-_23768234 | 0.25 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr10_-_32494499 | 0.25 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr6_+_102506 | 0.25 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr1_-_46981134 | 0.25 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr8_-_12867434 | 0.25 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr19_+_390298 | 0.24 |
ENSDART00000136361
|
sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr17_-_40956035 | 0.20 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr4_-_20292821 | 0.19 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr17_-_41798856 | 0.19 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr7_+_48667081 | 0.19 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr19_-_32641725 | 0.18 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr4_-_9852318 | 0.18 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr7_+_2236317 | 0.18 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr22_-_8725768 | 0.17 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr10_+_6884123 | 0.17 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr18_+_50461981 | 0.17 |
ENSDART00000158761
|
CU896640.1
|
|
| chr8_+_25034544 | 0.17 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr14_-_8940499 | 0.17 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr4_+_2655358 | 0.16 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr20_-_45060241 | 0.16 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr24_-_25004553 | 0.15 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr11_+_18873619 | 0.15 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr11_-_44801968 | 0.14 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr4_-_71912457 | 0.12 |
ENSDART00000179685
|
si:dkey-92c21.1
|
si:dkey-92c21.1 |
| chr12_+_30367371 | 0.12 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr18_+_2228737 | 0.11 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr11_-_45138857 | 0.11 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr22_-_881725 | 0.10 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr19_-_30524952 | 0.10 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr21_-_35419486 | 0.09 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr24_+_12894282 | 0.08 |
ENSDART00000061301
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CR354582.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.5 | 1.4 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.4 | 1.8 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 4.0 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.4 | 1.3 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.4 | 2.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.3 | 1.6 | GO:0022410 | circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) |
| 0.2 | 1.3 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 2.0 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 2.7 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 0.7 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 1.5 | GO:0042044 | fluid transport(GO:0042044) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.7 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 0.5 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 1.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 2.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 4.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.4 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 1.7 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 2.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 1.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.9 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 1.2 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 2.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.7 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 9.4 | GO:0050953 | sensory perception of light stimulus(GO:0050953) |
| 0.0 | 1.2 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.7 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 1.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 2.3 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 2.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.8 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.5 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.0 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.4 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 1.2 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 1.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0070257 | secretion by tissue(GO:0032941) mucus secretion(GO:0070254) regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 5.7 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 0.7 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.8 | 6.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.3 | 1.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 1.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 2.0 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 1.0 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 3.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.4 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 2.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.7 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 2.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.0 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.9 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 2.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.3 | 1.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.3 | 2.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 6.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 2.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.9 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 2.0 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 3.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.7 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.6 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 4.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 2.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.1 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID EPO PATHWAY | EPO signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |