Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
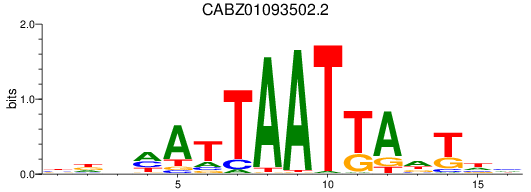
Results for CABZ01093502.2
Z-value: 0.50
Transcription factors associated with CABZ01093502.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CABZ01093502.2
|
ENSDARG00000105053 | developing brain homeobox 2 |
Activity profile of CABZ01093502.2 motif
Sorted Z-values of CABZ01093502.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_16485190 | 1.21 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr16_+_54209504 | 1.11 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr20_+_11731039 | 1.09 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr21_+_28478663 | 0.95 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr3_-_19368435 | 0.89 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr17_+_8799661 | 0.86 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr6_-_58764672 | 0.77 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr8_-_30204650 | 0.76 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr20_+_40457599 | 0.73 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr6_+_7533601 | 0.72 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr22_-_24818066 | 0.70 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr6_-_35046735 | 0.70 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr17_-_16422654 | 0.68 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr22_-_24791505 | 0.66 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr2_-_10877765 | 0.66 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr21_-_20328375 | 0.65 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr21_+_43172506 | 0.60 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr8_-_50888806 | 0.59 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr20_+_25586099 | 0.59 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr17_-_33412868 | 0.56 |
ENSDART00000187521
|
BX323819.1
|
|
| chr22_-_36530902 | 0.56 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr21_+_5926777 | 0.55 |
ENSDART00000121769
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr2_+_10878406 | 0.52 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr24_-_38657683 | 0.52 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr5_+_13394543 | 0.51 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr11_+_17984167 | 0.49 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr8_+_50953776 | 0.48 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr12_-_28363111 | 0.48 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr24_+_28953089 | 0.46 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr11_-_1509773 | 0.46 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr13_-_12602920 | 0.42 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr15_-_43284021 | 0.41 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr17_+_20589553 | 0.37 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr3_+_25166805 | 0.37 |
ENSDART00000077493
|
TST
|
zgc:162544 |
| chr22_-_14247276 | 0.35 |
ENSDART00000033332
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr25_+_3318192 | 0.34 |
ENSDART00000146154
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr9_+_21793565 | 0.31 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr6_+_12527725 | 0.30 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr17_-_12764360 | 0.30 |
ENSDART00000003418
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr18_+_30847237 | 0.29 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr23_+_4709607 | 0.29 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr23_-_6522099 | 0.28 |
ENSDART00000092214
ENSDART00000183380 ENSDART00000138020 |
bmp7b
|
bone morphogenetic protein 7b |
| chr2_+_55984788 | 0.27 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr2_-_7845110 | 0.27 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr5_+_33498253 | 0.26 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr7_-_17412559 | 0.25 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr5_+_34623107 | 0.25 |
ENSDART00000184126
|
enc1
|
ectodermal-neural cortex 1 |
| chr6_+_13117598 | 0.24 |
ENSDART00000104744
|
casp8l1
|
caspase 8, apoptosis-related cysteine peptidase, like 1 |
| chr17_-_10122204 | 0.23 |
ENSDART00000160751
|
BX088587.1
|
|
| chr3_-_40054615 | 0.23 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr13_+_35528607 | 0.21 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr16_+_23303859 | 0.21 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr2_+_19522082 | 0.21 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr16_+_24733741 | 0.20 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr5_+_9224051 | 0.17 |
ENSDART00000139265
|
si:ch211-12e13.12
|
si:ch211-12e13.12 |
| chr2_-_59327299 | 0.16 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr1_+_58242498 | 0.15 |
ENSDART00000149091
|
ggt1l2.2
|
gamma-glutamyltransferase 1 like 2.2 |
| chr23_+_21405201 | 0.15 |
ENSDART00000144409
|
iffo2a
|
intermediate filament family orphan 2a |
| chr4_+_59845617 | 0.15 |
ENSDART00000167626
ENSDART00000123157 |
si:dkey-196n19.2
|
si:dkey-196n19.2 |
| chr8_-_53044300 | 0.14 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr12_-_10409961 | 0.14 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr2_+_19578446 | 0.13 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr19_+_43780970 | 0.13 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr22_+_2751887 | 0.12 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr3_+_26145013 | 0.11 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr19_+_33093577 | 0.09 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr15_-_21014270 | 0.09 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr8_-_20230802 | 0.08 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr2_+_14992879 | 0.07 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr9_-_32177117 | 0.07 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr13_-_38730267 | 0.07 |
ENSDART00000157524
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr10_-_20339113 | 0.06 |
ENSDART00000144378
ENSDART00000098600 |
zmat4b
|
zinc finger, matrin-type 4b |
| chr20_-_9095105 | 0.05 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr9_-_51436377 | 0.05 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr23_+_384850 | 0.04 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr1_-_31171242 | 0.00 |
ENSDART00000190294
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01093502.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.8 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.7 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.5 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.9 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 1.4 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0090076 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.6 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 1.4 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.2 | 0.8 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.7 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 1.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |