Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for CABZ01066694.1
Z-value: 0.48
Transcription factors associated with CABZ01066694.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CABZ01066694.1
|
ENSDARG00000110178 | ENSDARG00000110178 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CABZ01066694.1 | dr11_v1_chr6_+_168962_168962 | 0.55 | 1.5e-02 | Click! |
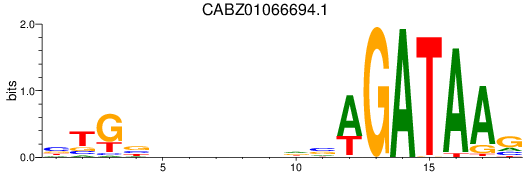
Activity profile of CABZ01066694.1 motif
Sorted Z-values of CABZ01066694.1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_6109861 | 2.10 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr19_+_37509638 | 2.04 |
ENSDART00000139999
|
si:ch211-250g4.3
|
si:ch211-250g4.3 |
| chr7_-_38638809 | 1.82 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr2_-_54054225 | 1.59 |
ENSDART00000167239
|
CABZ01050249.1
|
|
| chr9_-_22205682 | 1.58 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr20_+_33981946 | 1.28 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr1_-_58766424 | 1.24 |
ENSDART00000191010
|
CABZ01096573.1
|
|
| chr6_+_41038757 | 1.17 |
ENSDART00000011245
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr2_-_32768951 | 1.12 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr2_-_56635744 | 0.85 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr9_-_52814204 | 0.85 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr15_+_9294620 | 0.78 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr15_-_33925851 | 0.78 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr18_-_21188768 | 0.75 |
ENSDART00000060166
|
got2a
|
glutamic-oxaloacetic transaminase 2a, mitochondrial |
| chr7_+_26138240 | 0.73 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr21_-_22547496 | 0.72 |
ENSDART00000166835
ENSDART00000089030 |
myo5b
|
myosin VB |
| chr25_-_16742438 | 0.70 |
ENSDART00000156091
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr16_-_30931651 | 0.70 |
ENSDART00000132643
|
slc45a4
|
solute carrier family 45, member 4 |
| chr10_+_21677058 | 0.69 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr25_-_26018424 | 0.67 |
ENSDART00000089332
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr25_-_26018240 | 0.66 |
ENSDART00000150800
|
acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr16_+_29630965 | 0.64 |
ENSDART00000185820
ENSDART00000192105 ENSDART00000153683 ENSDART00000186713 |
VPS72
|
si:ch211-203d17.1 |
| chr17_-_23609210 | 0.63 |
ENSDART00000064003
|
ifit15
|
interferon-induced protein with tetratricopeptide repeats 15 |
| chr7_+_8324506 | 0.61 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr20_-_51727860 | 0.59 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr10_-_32558917 | 0.58 |
ENSDART00000128888
ENSDART00000143301 |
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr10_+_17936441 | 0.58 |
ENSDART00000146489
ENSDART00000064866 |
prkab1a
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, a |
| chr25_-_32311048 | 0.57 |
ENSDART00000181806
ENSDART00000086334 |
CU372926.1
|
|
| chr3_+_52737565 | 0.57 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr9_+_55379283 | 0.56 |
ENSDART00000192728
|
nlgn4b
|
neuroligin 4b |
| chr15_+_45563656 | 0.55 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr4_+_76692834 | 0.55 |
ENSDART00000064315
ENSDART00000137653 |
ms4a17a.1
|
membrane-spanning 4-domains, subfamily A, member 17A.1 |
| chr8_-_7637626 | 0.54 |
ENSDART00000123882
ENSDART00000040672 |
mecp2
|
methyl CpG binding protein 2 |
| chr9_-_42871756 | 0.54 |
ENSDART00000191396
|
ttn.1
|
titin, tandem duplicate 1 |
| chr5_-_7897316 | 0.53 |
ENSDART00000160743
ENSDART00000160912 |
opn4xb
|
opsin 4xb |
| chr16_+_41067586 | 0.52 |
ENSDART00000181876
|
scap
|
SREBF chaperone |
| chr14_+_17197132 | 0.50 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr3_-_31019715 | 0.49 |
ENSDART00000156687
ENSDART00000156127 ENSDART00000153945 |
si:dkeyp-71f10.5
|
si:dkeyp-71f10.5 |
| chr7_+_1521834 | 0.48 |
ENSDART00000174007
|
si:cabz01102082.1
|
si:cabz01102082.1 |
| chr8_+_28900689 | 0.48 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr16_+_45930962 | 0.47 |
ENSDART00000124689
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr4_+_69619 | 0.47 |
ENSDART00000164425
|
mansc1
|
MANSC domain containing 1 |
| chr16_-_45069882 | 0.47 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr22_+_8551813 | 0.46 |
ENSDART00000129690
|
zmp:0000000984
|
zmp:0000000984 |
| chr14_+_21783229 | 0.46 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr25_-_32464149 | 0.46 |
ENSDART00000152787
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr7_+_34305903 | 0.43 |
ENSDART00000173575
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr15_+_45563491 | 0.42 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr7_+_9922607 | 0.41 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr3_+_21669545 | 0.40 |
ENSDART00000156527
|
crhr1
|
corticotropin releasing hormone receptor 1 |
| chr24_-_21498802 | 0.40 |
ENSDART00000181235
ENSDART00000153695 |
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr4_-_75681004 | 0.39 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr7_-_68419830 | 0.37 |
ENSDART00000191606
|
CU468723.2
|
|
| chr6_+_58280936 | 0.37 |
ENSDART00000155244
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr10_+_21584195 | 0.37 |
ENSDART00000100599
|
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr20_+_27194833 | 0.36 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr20_-_22378401 | 0.35 |
ENSDART00000011135
ENSDART00000110967 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr2_-_43850637 | 0.35 |
ENSDART00000136077
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr7_+_41748693 | 0.35 |
ENSDART00000174379
ENSDART00000052168 |
hrh3
|
histamine receptor H3 |
| chr24_-_26622423 | 0.35 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr15_+_34592215 | 0.34 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr8_+_31872992 | 0.34 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr10_+_15255012 | 0.32 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr13_+_46718518 | 0.31 |
ENSDART00000160401
ENSDART00000182884 |
tmem63ba
|
transmembrane protein 63Ba |
| chr25_-_21550222 | 0.31 |
ENSDART00000185404
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr14_+_21783400 | 0.30 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr7_-_50883433 | 0.30 |
ENSDART00000174314
|
PAQR9
|
progestin and adipoQ receptor family member 9 |
| chr21_+_198269 | 0.30 |
ENSDART00000193485
ENSDART00000163696 |
gsdf
|
gonadal somatic cell derived factor |
| chr3_+_17346502 | 0.30 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr6_-_49078263 | 0.30 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr17_+_44780166 | 0.29 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr11_+_14937904 | 0.29 |
ENSDART00000185103
|
CR550302.3
|
|
| chr25_+_6186823 | 0.28 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr5_+_19165949 | 0.28 |
ENSDART00000140710
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr3_-_11203689 | 0.28 |
ENSDART00000166328
|
BX323882.1
|
|
| chr2_+_49713592 | 0.27 |
ENSDART00000189624
|
BX323861.3
|
|
| chr2_+_8779164 | 0.27 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr21_-_22648007 | 0.27 |
ENSDART00000121788
|
gig2l
|
grass carp reovirus (GCRV)-induced gene 2l |
| chr18_+_45228691 | 0.27 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr1_+_9082417 | 0.27 |
ENSDART00000146303
|
cacng3a
|
calcium channel, voltage-dependent, gamma subunit 3a |
| chr25_-_12805295 | 0.27 |
ENSDART00000157629
|
ca5a
|
carbonic anhydrase Va |
| chr10_+_15255198 | 0.27 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr8_+_7778770 | 0.26 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr6_-_32349153 | 0.26 |
ENSDART00000140004
|
angptl3
|
angiopoietin-like 3 |
| chr20_+_38322444 | 0.26 |
ENSDART00000161741
ENSDART00000132241 ENSDART00000148936 ENSDART00000149623 |
stum
|
stum, mechanosensory transduction mediator homolog |
| chr6_-_49078702 | 0.26 |
ENSDART00000135185
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr25_+_16098620 | 0.25 |
ENSDART00000142564
ENSDART00000165598 |
far1
|
fatty acyl CoA reductase 1 |
| chr2_-_43850897 | 0.24 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr3_-_58205045 | 0.24 |
ENSDART00000130740
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr4_-_22338906 | 0.24 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr12_+_7497882 | 0.24 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr14_+_46268248 | 0.23 |
ENSDART00000172979
|
cabp2b
|
calcium binding protein 2b |
| chr9_-_2936017 | 0.21 |
ENSDART00000102823
|
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr22_+_6254194 | 0.21 |
ENSDART00000112388
ENSDART00000135176 |
rnasel4
|
ribonuclease like 4 |
| chr24_-_39858710 | 0.20 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr15_-_37719015 | 0.19 |
ENSDART00000154875
|
si:dkey-117a8.1
|
si:dkey-117a8.1 |
| chr10_+_38643304 | 0.18 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr15_-_14248047 | 0.18 |
ENSDART00000164762
|
si:ch211-11n16.3
|
si:ch211-11n16.3 |
| chr13_-_23031803 | 0.18 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr22_+_7923713 | 0.18 |
ENSDART00000167210
|
CABZ01034691.1
|
|
| chr16_+_54829293 | 0.17 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr12_-_20373058 | 0.16 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr2_+_16160906 | 0.16 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr16_-_50157187 | 0.16 |
ENSDART00000155162
|
nek10
|
NIMA-related kinase 10 |
| chr10_+_39476432 | 0.16 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr24_-_11050727 | 0.16 |
ENSDART00000145466
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr9_+_41156287 | 0.16 |
ENSDART00000189195
ENSDART00000186270 |
stat4
|
signal transducer and activator of transcription 4 |
| chr4_+_37936249 | 0.15 |
ENSDART00000184022
|
si:dkeyp-82b4.2
|
si:dkeyp-82b4.2 |
| chr25_+_2415131 | 0.15 |
ENSDART00000162210
|
zmp:0000000932
|
zmp:0000000932 |
| chr17_+_45737992 | 0.15 |
ENSDART00000135073
ENSDART00000143525 ENSDART00000158165 ENSDART00000184167 ENSDART00000109525 |
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr3_+_32789605 | 0.15 |
ENSDART00000171895
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr2_-_35103069 | 0.14 |
ENSDART00000111730
|
pappa2
|
pappalysin 2 |
| chr4_+_39742119 | 0.14 |
ENSDART00000176004
|
CR749167.2
|
|
| chr9_+_29585943 | 0.14 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr8_-_46045172 | 0.13 |
ENSDART00000135138
|
si:ch211-119d14.3
|
si:ch211-119d14.3 |
| chr5_-_68244564 | 0.13 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr5_-_67241633 | 0.12 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr10_-_10969596 | 0.12 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr16_-_42303856 | 0.12 |
ENSDART00000180030
|
ppox
|
protoporphyrinogen oxidase |
| chr17_+_23994633 | 0.12 |
ENSDART00000156988
|
si:ch211-63b16.3
|
si:ch211-63b16.3 |
| chr5_+_42388464 | 0.10 |
ENSDART00000191596
|
BX548073.13
|
|
| chr5_+_38200807 | 0.10 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr2_+_38742338 | 0.10 |
ENSDART00000177290
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr7_-_5191797 | 0.10 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr20_+_14968031 | 0.10 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
| chr13_+_30169681 | 0.10 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr15_-_21837207 | 0.09 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr19_+_38167468 | 0.09 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr6_-_36182115 | 0.08 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr4_+_59245846 | 0.08 |
ENSDART00000104921
|
si:dkey-105i14.1
|
si:dkey-105i14.1 |
| chr1_-_6028876 | 0.08 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr18_-_39473055 | 0.08 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr25_-_13320986 | 0.07 |
ENSDART00000169238
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr3_+_33341640 | 0.07 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr12_+_4971515 | 0.07 |
ENSDART00000161076
|
arhgap27
|
Rho GTPase activating protein 27 |
| chr9_+_41156818 | 0.07 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr7_+_5052336 | 0.06 |
ENSDART00000133393
|
si:ch211-272h9.6
|
si:ch211-272h9.6 |
| chr2_-_2020044 | 0.06 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr14_-_10371638 | 0.06 |
ENSDART00000003879
|
fam199x
|
family with sequence similarity 199, X-linked |
| chr18_+_34478959 | 0.05 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr11_+_25101220 | 0.05 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr5_-_30088070 | 0.05 |
ENSDART00000078115
ENSDART00000134209 |
sdhda
|
succinate dehydrogenase complex, subunit D, integral membrane protein a |
| chr6_-_42336987 | 0.04 |
ENSDART00000128777
ENSDART00000075601 |
fancd2
|
Fanconi anemia, complementation group D2 |
| chr1_-_26023678 | 0.04 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr13_+_42011287 | 0.04 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr5_-_37209481 | 0.03 |
ENSDART00000158614
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr15_+_17621375 | 0.03 |
ENSDART00000158352
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr16_-_24668620 | 0.03 |
ENSDART00000012807
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr11_-_45185792 | 0.03 |
ENSDART00000171328
|
si:dkey-93h22.7
|
si:dkey-93h22.7 |
| chr17_+_8175998 | 0.03 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr21_+_34814444 | 0.02 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr21_-_40040178 | 0.01 |
ENSDART00000159741
|
BX248108.1
|
Danio rerio multidrug and toxin extrusion protein 1-like (mate4), mRNA. |
| chr6_-_16394528 | 0.01 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr21_+_25935845 | 0.01 |
ENSDART00000141149
|
caln2
|
calneuron 2 |
| chr14_-_6943934 | 0.01 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr23_+_4689626 | 0.00 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
Network of associatons between targets according to the STRING database.
First level regulatory network of CABZ01066694.1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.2 | 0.8 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 0.7 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 0.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 0.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.6 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.5 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.5 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.5 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.9 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.1 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.1 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.3 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 1.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 0.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 0.8 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.6 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 1.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.3 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |