Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
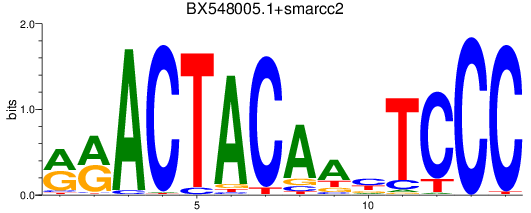
Results for BX548005.1+smarcc2
Z-value: 1.49
Transcription factors associated with BX548005.1+smarcc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smarcc2
|
ENSDARG00000077946 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
|
BX548005.1
|
ENSDARG00000110907 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smarcc2 | dr11_v1_chr6_+_39836474_39836474 | -0.91 | 8.4e-08 | Click! |
Activity profile of BX548005.1+smarcc2 motif
Sorted Z-values of BX548005.1+smarcc2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_2725594 | 5.10 |
ENSDART00000152120
|
akirin2
|
akirin 2 |
| chr15_+_2857556 | 4.00 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr20_-_2725930 | 3.59 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr11_+_29537756 | 3.56 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr1_+_9153141 | 3.53 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr13_+_24402406 | 3.49 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr24_+_29352039 | 3.18 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr16_+_27442549 | 3.11 |
ENSDART00000015688
|
invs
|
inversin |
| chr3_-_26184018 | 3.07 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr11_-_16021424 | 3.06 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr3_-_26183699 | 3.01 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr19_-_41518922 | 2.97 |
ENSDART00000164483
ENSDART00000062080 |
chrac1
|
chromatin accessibility complex 1 |
| chr7_+_2455344 | 2.91 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr25_-_3347418 | 2.87 |
ENSDART00000082385
|
golt1bb
|
golgi transport 1Bb |
| chr1_+_24557414 | 2.81 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr11_-_35171162 | 2.80 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr22_+_22302614 | 2.75 |
ENSDART00000049434
|
scamp4
|
secretory carrier membrane protein 4 |
| chr11_-_35171768 | 2.73 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr1_+_41131481 | 2.73 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr24_-_37680917 | 2.72 |
ENSDART00000131342
|
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr8_+_26410539 | 2.71 |
ENSDART00000168780
|
ifrd2
|
interferon-related developmental regulator 2 |
| chr1_+_5485799 | 2.64 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr17_-_12764360 | 2.58 |
ENSDART00000003418
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr24_+_21092156 | 2.51 |
ENSDART00000028542
|
ccdc191
|
coiled-coil domain containing 191 |
| chr14_+_989733 | 2.51 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr9_+_37754845 | 2.47 |
ENSDART00000100592
|
pdia5
|
protein disulfide isomerase family A, member 5 |
| chr2_-_25140022 | 2.43 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr16_-_41535690 | 2.40 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr19_-_17210760 | 2.40 |
ENSDART00000007906
|
stmn1a
|
stathmin 1a |
| chr18_-_27316599 | 2.40 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr17_-_18797245 | 2.35 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr8_+_48942470 | 2.29 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr3_+_31058464 | 2.28 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr8_-_16725959 | 2.28 |
ENSDART00000183593
|
depdc1a
|
DEP domain containing 1a |
| chr18_-_18587745 | 2.27 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr6_-_39700965 | 2.25 |
ENSDART00000156645
|
espl1
|
extra spindle pole bodies like 1, separase |
| chr9_-_23242684 | 2.20 |
ENSDART00000053282
ENSDART00000179770 |
ccnt2a
|
cyclin T2a |
| chr13_+_31390313 | 2.16 |
ENSDART00000111477
ENSDART00000137291 |
METTL18
|
si:dkey-15f17.8 |
| chr8_-_36554675 | 2.15 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr25_+_3347461 | 2.15 |
ENSDART00000104888
|
slc35b4
|
solute carrier family 35, member B4 |
| chr1_+_53919110 | 2.14 |
ENSDART00000020680
|
nup133
|
nucleoporin 133 |
| chr20_-_28842524 | 2.12 |
ENSDART00000046035
ENSDART00000139843 ENSDART00000129858 ENSDART00000137425 ENSDART00000135720 |
max
|
myc associated factor X |
| chr8_-_18613948 | 2.12 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr16_-_27442344 | 2.11 |
ENSDART00000027545
|
erp44
|
endoplasmic reticulum protein 44 |
| chr18_-_38245062 | 2.10 |
ENSDART00000189092
|
nat10
|
N-acetyltransferase 10 |
| chr25_+_9013342 | 2.09 |
ENSDART00000154207
ENSDART00000153705 |
im:7145024
|
im:7145024 |
| chr13_+_28785814 | 2.07 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr6_-_58975010 | 2.07 |
ENSDART00000144911
ENSDART00000144514 |
mars
|
methionyl-tRNA synthetase |
| chr16_-_39267185 | 2.06 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr23_+_5104743 | 2.04 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr18_-_38244871 | 2.03 |
ENSDART00000076399
|
nat10
|
N-acetyltransferase 10 |
| chr19_-_17210928 | 2.02 |
ENSDART00000164683
|
stmn1a
|
stathmin 1a |
| chr8_+_48943009 | 1.98 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr18_+_5308392 | 1.97 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr15_-_44052927 | 1.96 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr9_-_29579908 | 1.94 |
ENSDART00000140876
|
cenpj
|
centromere protein J |
| chr4_+_9592486 | 1.91 |
ENSDART00000080829
|
hspa14
|
heat shock protein 14 |
| chr18_+_29898955 | 1.91 |
ENSDART00000064080
|
cenpn
|
centromere protein N |
| chr14_-_12106603 | 1.85 |
ENSDART00000054619
|
prps1b
|
phosphoribosyl pyrophosphate synthetase 1B |
| chr22_+_25088999 | 1.83 |
ENSDART00000158225
|
rrbp1b
|
ribosome binding protein 1b |
| chr9_+_30478768 | 1.80 |
ENSDART00000101097
|
acp6
|
acid phosphatase 6, lysophosphatidic |
| chr7_-_26627252 | 1.80 |
ENSDART00000164050
ENSDART00000159826 |
phf23b
|
PHD finger protein 23b |
| chr15_-_34001000 | 1.79 |
ENSDART00000180464
|
VWDE
|
si:dkey-30e9.7 |
| chr17_+_43659940 | 1.79 |
ENSDART00000145738
ENSDART00000075619 |
adob
|
2-aminoethanethiol (cysteamine) dioxygenase b |
| chr6_+_8652310 | 1.79 |
ENSDART00000105098
|
usp40
|
ubiquitin specific peptidase 40 |
| chr23_+_2421313 | 1.79 |
ENSDART00000126038
|
tcp1
|
t-complex 1 |
| chr3_+_12554801 | 1.75 |
ENSDART00000167177
|
ccnf
|
cyclin F |
| chr21_+_8198652 | 1.74 |
ENSDART00000011096
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr6_-_50730749 | 1.74 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr5_-_10082244 | 1.73 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr5_-_26170225 | 1.73 |
ENSDART00000146363
ENSDART00000017696 |
fam151b
|
family with sequence similarity 151, member B |
| chr9_+_56232548 | 1.72 |
ENSDART00000099276
|
cnot11
|
CCR4-NOT transcription complex, subunit 11 |
| chr14_-_4321874 | 1.71 |
ENSDART00000042672
|
guf1
|
GUF1 homolog, GTPase |
| chr4_-_14954029 | 1.70 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr3_+_24060454 | 1.69 |
ENSDART00000143088
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr2_+_27394798 | 1.67 |
ENSDART00000115071
|
selenop2
|
selenoprotein P2 |
| chr8_-_48847772 | 1.67 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr21_-_26071773 | 1.65 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr22_-_3152357 | 1.65 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr15_+_37589698 | 1.63 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr16_+_9762261 | 1.63 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr23_+_2421689 | 1.62 |
ENSDART00000180200
|
tcp1
|
t-complex 1 |
| chr15_+_8767650 | 1.61 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr19_+_15521997 | 1.61 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr23_+_20640484 | 1.61 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr2_+_27394979 | 1.59 |
ENSDART00000170495
|
selenop2
|
selenoprotein P2 |
| chr18_-_16953978 | 1.59 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr8_+_12951155 | 1.58 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr19_+_26072624 | 1.57 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr10_+_8091144 | 1.56 |
ENSDART00000143848
ENSDART00000075485 |
sub1a
|
SUB1 homolog, transcriptional regulator a |
| chr7_+_71683853 | 1.55 |
ENSDART00000163002
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr16_-_55259199 | 1.55 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr11_-_33868881 | 1.54 |
ENSDART00000163295
ENSDART00000172633 ENSDART00000171439 |
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr5_-_67349916 | 1.54 |
ENSDART00000144092
|
mlxip
|
MLX interacting protein |
| chr25_-_8625601 | 1.54 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr3_+_27665160 | 1.53 |
ENSDART00000103660
|
clcn7
|
chloride channel 7 |
| chr21_+_13327527 | 1.53 |
ENSDART00000114294
|
snrpd3l
|
small nuclear ribonucleoprotein D3 polypeptide, like |
| chr3_+_36646054 | 1.53 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr1_-_51720633 | 1.51 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr20_+_5985329 | 1.48 |
ENSDART00000165489
|
cep128
|
centrosomal protein 128 |
| chr25_+_20715950 | 1.46 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr4_+_17327704 | 1.46 |
ENSDART00000016075
ENSDART00000133160 |
nup37
|
nucleoporin 37 |
| chr5_-_41142467 | 1.45 |
ENSDART00000129415
|
zfr
|
zinc finger RNA binding protein |
| chr13_-_24745288 | 1.44 |
ENSDART00000031564
|
sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
| chr20_-_3997531 | 1.44 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr24_+_42004640 | 1.43 |
ENSDART00000171380
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr20_-_31743817 | 1.41 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr14_+_30774894 | 1.40 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr23_-_20133994 | 1.39 |
ENSDART00000004871
|
lrrc23
|
leucine rich repeat containing 23 |
| chr8_+_49936585 | 1.39 |
ENSDART00000098707
|
naa35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr20_-_16849306 | 1.38 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr4_-_14954327 | 1.38 |
ENSDART00000182729
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr22_-_24285432 | 1.38 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr10_-_44306636 | 1.37 |
ENSDART00000191068
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr1_+_35494837 | 1.36 |
ENSDART00000140724
|
gab1
|
GRB2-associated binding protein 1 |
| chr18_-_46369516 | 1.35 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr8_+_31016180 | 1.34 |
ENSDART00000130870
ENSDART00000143604 |
odf2b
|
outer dense fiber of sperm tails 2b |
| chr13_+_39315881 | 1.34 |
ENSDART00000135999
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr13_-_15929402 | 1.34 |
ENSDART00000090273
|
ttl
|
tubulin tyrosine ligase |
| chr2_+_16710889 | 1.33 |
ENSDART00000017852
|
ubxn7
|
UBX domain protein 7 |
| chr14_-_26411918 | 1.33 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr8_-_36618073 | 1.31 |
ENSDART00000047912
|
gpkow
|
G patch domain and KOW motifs |
| chr6_-_6993046 | 1.31 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr19_-_8536474 | 1.31 |
ENSDART00000139715
|
dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr19_+_627899 | 1.29 |
ENSDART00000148508
|
tert
|
telomerase reverse transcriptase |
| chr12_+_8474868 | 1.28 |
ENSDART00000062858
|
adoa
|
2-aminoethanethiol (cysteamine) dioxygenase a |
| chr22_-_506522 | 1.28 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr21_-_21020708 | 1.27 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr22_+_22888 | 1.26 |
ENSDART00000082471
|
mfap2
|
microfibril associated protein 2 |
| chr2_+_36608387 | 1.26 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr9_-_29985390 | 1.25 |
ENSDART00000134157
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr5_+_15495351 | 1.25 |
ENSDART00000111646
ENSDART00000114446 |
suds3
|
SDS3 homolog, SIN3A corepressor complex component |
| chr20_+_14789305 | 1.25 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr13_+_33655404 | 1.24 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr16_+_31921812 | 1.24 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr3_+_3681116 | 1.24 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr4_-_22363709 | 1.23 |
ENSDART00000037670
|
orc5
|
origin recognition complex, subunit 5 |
| chr7_+_7696665 | 1.22 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr8_+_23381892 | 1.22 |
ENSDART00000180950
ENSDART00000063010 ENSDART00000074241 ENSDART00000142783 |
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr21_+_25802190 | 1.22 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr22_+_23546926 | 1.21 |
ENSDART00000157940
|
aspm
|
abnormal spindle microtubule assembly |
| chr16_+_46684855 | 1.20 |
ENSDART00000058325
|
lamtor2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr18_+_46162204 | 1.18 |
ENSDART00000113545
ENSDART00000147556 |
zgc:113340
|
zgc:113340 |
| chr9_+_29985010 | 1.18 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr18_-_3166726 | 1.17 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr12_-_28363111 | 1.16 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr13_-_32626247 | 1.15 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr20_-_36809059 | 1.14 |
ENSDART00000062925
|
slc25a27
|
solute carrier family 25, member 27 |
| chr2_+_31308587 | 1.14 |
ENSDART00000027090
|
clul1
|
clusterin-like 1 (retinal) |
| chr5_-_41142768 | 1.13 |
ENSDART00000074789
|
zfr
|
zinc finger RNA binding protein |
| chr24_+_37709191 | 1.10 |
ENSDART00000066558
|
decr2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr11_+_16040517 | 1.10 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr7_-_38087865 | 1.09 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr2_-_1468258 | 1.09 |
ENSDART00000114431
|
gpaa1
|
glycosylphosphatidylinositol anchor attachment 1 |
| chr15_+_15516612 | 1.09 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr20_-_18313864 | 1.09 |
ENSDART00000015479
|
psma8
|
proteasome subunit alpha 8 |
| chr8_-_11988065 | 1.07 |
ENSDART00000005140
|
med27
|
mediator complex subunit 27 |
| chr15_-_23442891 | 1.07 |
ENSDART00000059376
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr3_+_1219344 | 1.06 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr15_-_29556757 | 1.06 |
ENSDART00000060049
|
hspa13
|
heat shock protein 70 family, member 13 |
| chr3_+_27664864 | 1.04 |
ENSDART00000126533
ENSDART00000180848 |
clcn7
|
chloride channel 7 |
| chr19_+_48018802 | 1.04 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr16_+_31922065 | 1.04 |
ENSDART00000131661
ENSDART00000144194 ENSDART00000145510 |
rps9
|
ribosomal protein S9 |
| chr16_-_27161410 | 1.03 |
ENSDART00000177503
|
LO017771.1
|
|
| chr25_+_20716176 | 1.03 |
ENSDART00000073651
|
ergic2
|
ERGIC and golgi 2 |
| chr23_+_33947874 | 1.03 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr5_-_32890807 | 1.02 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr23_-_14216506 | 1.02 |
ENSDART00000019620
|
ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr23_-_26227805 | 1.02 |
ENSDART00000158082
|
BX927204.1
|
|
| chr16_+_30604387 | 1.01 |
ENSDART00000058785
|
fam210ab
|
family with sequence similarity 210, member Ab |
| chr4_-_1818315 | 1.00 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr7_-_17712665 | 1.00 |
ENSDART00000149047
|
men1
|
multiple endocrine neoplasia I |
| chr21_+_21791799 | 1.00 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr13_-_33654931 | 0.99 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr4_+_25912654 | 0.99 |
ENSDART00000109508
ENSDART00000134218 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr18_+_8320165 | 0.98 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr5_+_60590796 | 0.97 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr10_+_36441124 | 0.95 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr16_-_34285106 | 0.95 |
ENSDART00000044235
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr17_-_24521382 | 0.94 |
ENSDART00000092948
|
peli1b
|
pellino E3 ubiquitin protein ligase 1b |
| chr24_+_19591893 | 0.93 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr5_-_56948058 | 0.93 |
ENSDART00000083074
ENSDART00000191028 |
si:ch211-127d4.3
|
si:ch211-127d4.3 |
| chr20_+_9474841 | 0.93 |
ENSDART00000053847
|
rad51b
|
RAD51 paralog B |
| chr13_-_33639050 | 0.93 |
ENSDART00000133073
|
rrbp1a
|
ribosome binding protein 1a |
| chr2_-_16159491 | 0.93 |
ENSDART00000110059
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr16_+_42471455 | 0.92 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr15_+_44053244 | 0.92 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr23_+_33752275 | 0.92 |
ENSDART00000007260
|
si:ch211-210c8.6
|
si:ch211-210c8.6 |
| chr3_+_4502066 | 0.91 |
ENSDART00000088610
|
rangap1a
|
RAN GTPase activating protein 1a |
| chr2_-_13254821 | 0.91 |
ENSDART00000022621
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr20_+_14789148 | 0.91 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr15_-_37589600 | 0.91 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr12_+_36109507 | 0.91 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr25_+_36311333 | 0.90 |
ENSDART00000190174
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr13_-_42400647 | 0.89 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr7_-_24022340 | 0.88 |
ENSDART00000149133
|
cideb
|
cell death-inducing DFFA-like effector b |
| chr11_+_18612421 | 0.87 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr4_+_25912308 | 0.86 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr11_+_34921492 | 0.86 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr6_-_34938678 | 0.86 |
ENSDART00000186689
ENSDART00000131610 |
serbp1a
|
SERPINE1 mRNA binding protein 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of BX548005.1+smarcc2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.8 | 3.2 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.8 | 3.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.8 | 3.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.6 | 4.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.6 | 2.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.5 | 2.7 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.5 | 3.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.5 | 2.1 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.5 | 1.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.5 | 2.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.5 | 4.0 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.5 | 2.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.5 | 1.9 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.4 | 1.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.4 | 1.8 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.4 | 1.3 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.4 | 1.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.3 | 3.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.3 | 1.8 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.3 | 2.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.3 | 1.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.3 | 2.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.3 | 1.9 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.3 | 1.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 0.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.3 | 2.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 1.0 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.3 | 1.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 2.2 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 | 1.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 0.6 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.2 | 3.0 | GO:0042026 | protein refolding(GO:0042026) |
| 0.2 | 1.4 | GO:0032475 | otolith formation(GO:0032475) |
| 0.2 | 1.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 1.0 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 0.9 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 2.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 2.4 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 0.7 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.8 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 1.2 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.2 | 1.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.1 | 0.4 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 1.6 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 2.9 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 | 1.3 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 1.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 4.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 2.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 0.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.8 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 8.7 | GO:0045089 | positive regulation of innate immune response(GO:0045089) |
| 0.1 | 1.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 2.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 3.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 1.0 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 2.2 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.1 | 5.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 4.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 2.2 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 1.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.6 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 0.9 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 1.3 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 1.8 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 1.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.9 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 3.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 2.1 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.1 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.5 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.2 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.1 | 2.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 0.3 | GO:0060055 | angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 2.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.4 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.1 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.5 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 1.6 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 3.5 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 1.1 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.1 | 0.6 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 1.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 1.7 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.6 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.6 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 1.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.8 | GO:0039021 | regulation of protein stability(GO:0031647) pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 2.8 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 1.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 2.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 1.5 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.7 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 1.8 | GO:1901653 | cellular response to peptide hormone stimulus(GO:0071375) cellular response to peptide(GO:1901653) |
| 0.0 | 2.6 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 3.0 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.2 | GO:2000765 | regulation of cytoplasmic translation(GO:2000765) negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0043200 | response to amino acid(GO:0043200) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.6 | GO:1903038 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 1.5 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.7 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.4 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.0 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 1.3 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.1 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 0.6 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.0 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.6 | 1.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.5 | 2.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.5 | 1.9 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.5 | 1.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.4 | 3.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 1.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.4 | 1.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.1 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 2.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 4.0 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.3 | 3.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 3.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 1.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.3 | 1.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 2.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 2.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.3 | 2.3 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.2 | 2.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 5.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 1.8 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.2 | 0.6 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.2 | 4.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.2 | 1.9 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 2.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 1.7 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.8 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 2.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.8 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 4.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 5.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 4.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.9 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 3.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.5 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 3.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 8.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 6.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.3 | GO:0044420 | extracellular matrix component(GO:0044420) |
| 0.0 | 1.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.7 | 3.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 3.2 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.5 | 2.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.5 | 2.8 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.5 | 1.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.5 | 2.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.4 | 2.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.4 | 1.3 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.4 | 1.3 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.4 | 1.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.3 | 2.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.3 | 1.6 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.3 | 1.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 0.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 2.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 1.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 2.0 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 1.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.8 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 3.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.2 | 1.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 2.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 1.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 1.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.2 | 0.6 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.2 | 1.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.5 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 1.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.1 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 2.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 3.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 5.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.8 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 4.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.9 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.3 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 2.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.0 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.5 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 3.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 4.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 12.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 1.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.0 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 4.1 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.0 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.2 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.3 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 1.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 2.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.0 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 1.1 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.3 | 1.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 4.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 3.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 2.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 1.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 5.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.2 | 3.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 1.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 2.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.7 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 2.0 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 4.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 2.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 1.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.7 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 1.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 2.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |