Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
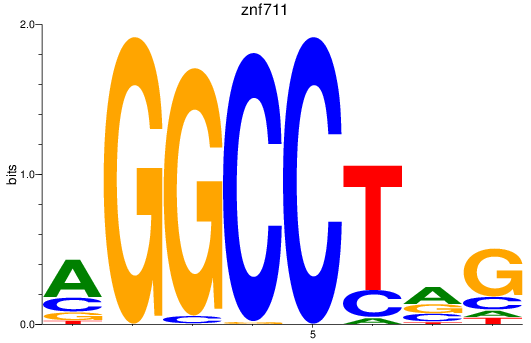
Results for znf711
Z-value: 1.37
Transcription factors associated with znf711
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf711
|
ENSDARG00000071868 | Danio rerio zinc finger protein 711 (znf711), transcript variant 1, mRNA. |
|
znf711
|
ENSDARG00000111963 | zinc finger protein 711 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf711 | dr11_v1_chr14_-_12020653_12020653 | -0.15 | 1.5e-01 | Click! |
Activity profile of znf711 motif
Sorted Z-values of znf711 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_23645810 | 24.07 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr5_+_51594209 | 22.74 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr23_-_10175898 | 19.88 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr13_+_22480496 | 17.90 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr3_+_32526799 | 12.86 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr3_+_23221047 | 12.32 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr13_+_24280380 | 11.45 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr7_+_7048245 | 10.92 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr8_-_1051438 | 10.89 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr13_+_22480857 | 10.81 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr13_+_22479988 | 10.00 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr17_-_15546862 | 9.99 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr7_+_29952719 | 9.35 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr17_-_10025234 | 7.70 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr23_-_10177442 | 7.56 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr11_-_55224 | 7.21 |
ENSDART00000159169
|
col2a1b
|
collagen, type II, alpha 1b |
| chr9_-_33107237 | 7.20 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr5_-_72390259 | 6.33 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr3_-_43770876 | 6.18 |
ENSDART00000160162
|
zgc:92162
|
zgc:92162 |
| chr23_+_13814978 | 6.06 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr19_+_31183495 | 5.99 |
ENSDART00000088618
|
meox2b
|
mesenchyme homeobox 2b |
| chr19_+_43119698 | 5.98 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr7_+_22747915 | 5.94 |
ENSDART00000173609
|
fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr11_-_22599584 | 5.93 |
ENSDART00000014062
|
myog
|
myogenin |
| chr7_+_22543963 | 5.92 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr2_+_25245083 | 5.79 |
ENSDART00000189649
ENSDART00000139080 |
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr11_+_11504014 | 5.69 |
ENSDART00000104264
ENSDART00000134806 ENSDART00000132291 |
zgc:171226
|
zgc:171226 |
| chr18_-_50839033 | 5.67 |
ENSDART00000169773
|
ddb1
|
damage-specific DNA binding protein 1 |
| chr11_-_3860534 | 5.27 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr6_+_28877306 | 5.17 |
ENSDART00000065137
ENSDART00000123189 ENSDART00000065135 ENSDART00000181512 ENSDART00000130799 |
tp63
|
tumor protein p63 |
| chr25_-_26736088 | 5.17 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr7_-_35432901 | 5.13 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr5_-_47727819 | 5.07 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr24_+_42074143 | 5.02 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr20_+_26538137 | 4.90 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr15_-_34567370 | 4.77 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr5_-_1963498 | 4.67 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr7_-_48667056 | 4.66 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr24_+_25471196 | 4.42 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr9_-_105135 | 4.27 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr16_-_24194587 | 4.17 |
ENSDART00000181520
|
rps19
|
ribosomal protein S19 |
| chr13_+_32148338 | 4.06 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr2_+_5371492 | 4.05 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr6_-_10835849 | 4.00 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr3_-_5644028 | 3.92 |
ENSDART00000019957
|
ddx39ab
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Ab |
| chr16_-_33817828 | 3.80 |
ENSDART00000188395
|
rspo1
|
R-spondin 1 |
| chr6_-_15096556 | 3.75 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr19_+_48018802 | 3.74 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr3_-_29925482 | 3.70 |
ENSDART00000151525
|
grna
|
granulin a |
| chr23_+_22819971 | 3.58 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr7_-_52531252 | 3.58 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr15_-_35960250 | 3.44 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr1_+_8508753 | 3.43 |
ENSDART00000091683
|
alkbh5
|
alkB homolog 5, RNA demethylase |
| chr10_-_38243579 | 3.37 |
ENSDART00000150159
|
usp25
|
ubiquitin specific peptidase 25 |
| chr20_+_1329509 | 3.37 |
ENSDART00000017791
ENSDART00000136669 |
tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr9_-_34260214 | 3.36 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr7_-_51639699 | 3.31 |
ENSDART00000128917
|
rps4x
|
ribosomal protein S4, X-linked |
| chr16_+_31921812 | 3.30 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr16_-_24195252 | 3.26 |
ENSDART00000136205
ENSDART00000048599 ENSDART00000161547 |
rps19
|
ribosomal protein S19 |
| chr10_+_26669177 | 3.22 |
ENSDART00000143402
|
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr25_+_3099073 | 3.15 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr1_-_54972170 | 3.12 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr15_-_36249087 | 3.04 |
ENSDART00000157190
|
ostn
|
osteocrin |
| chr9_+_33145522 | 3.03 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr21_-_45871866 | 2.93 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr8_+_25733872 | 2.89 |
ENSDART00000156340
|
si:ch211-167b20.8
|
si:ch211-167b20.8 |
| chr1_-_54971968 | 2.87 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr8_+_26372115 | 2.86 |
ENSDART00000053460
|
nat6
|
N-acetyltransferase 6 |
| chr13_+_21870269 | 2.82 |
ENSDART00000144612
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr20_+_33981946 | 2.76 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr16_+_27345383 | 2.72 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr5_+_45000597 | 2.72 |
ENSDART00000051146
|
dmrt3a
|
doublesex and mab-3 related transcription factor 3a |
| chr5_-_4418555 | 2.70 |
ENSDART00000170158
|
apooa
|
apolipoprotein O, a |
| chr6_+_58832323 | 2.69 |
ENSDART00000042595
|
dctn2
|
dynactin 2 (p50) |
| chr14_-_52408307 | 2.68 |
ENSDART00000167045
ENSDART00000162719 ENSDART00000160293 |
polr2b
|
polymerase (RNA) II (DNA directed) polypeptide B |
| chr3_+_60957512 | 2.64 |
ENSDART00000044096
|
helz
|
helicase with zinc finger |
| chr5_-_18007077 | 2.64 |
ENSDART00000129878
|
zdhhc8b
|
zinc finger, DHHC-type containing 8b |
| chr23_-_3758637 | 2.63 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr7_-_18508815 | 2.60 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr19_-_48019366 | 2.57 |
ENSDART00000169429
|
si:ch1073-205c8.5
|
si:ch1073-205c8.5 |
| chr7_+_40081630 | 2.56 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr12_+_22680115 | 2.55 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr23_+_34047413 | 2.53 |
ENSDART00000143933
ENSDART00000123925 ENSDART00000176139 |
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr22_-_817479 | 2.50 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr19_-_81851 | 2.48 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr5_-_15851953 | 2.47 |
ENSDART00000173101
|
si:dkey-1k23.3
|
si:dkey-1k23.3 |
| chr21_+_11969603 | 2.45 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr20_+_19176959 | 2.39 |
ENSDART00000163276
ENSDART00000020099 |
blk
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr23_-_20010579 | 2.36 |
ENSDART00000089999
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr19_-_7110617 | 2.35 |
ENSDART00000104838
|
psmb8a
|
proteasome subunit beta 8A |
| chr9_+_21407987 | 2.34 |
ENSDART00000145627
|
gja3
|
gap junction protein, alpha 3 |
| chr20_+_46492175 | 2.33 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr9_-_29036263 | 2.31 |
ENSDART00000160393
|
tmem177
|
transmembrane protein 177 |
| chr14_+_38849432 | 2.31 |
ENSDART00000052511
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr13_-_23007813 | 2.31 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr6_-_54107269 | 2.31 |
ENSDART00000190017
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr3_+_23752150 | 2.30 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr5_-_29238889 | 2.29 |
ENSDART00000143098
|
si:dkey-61l1.4
|
si:dkey-61l1.4 |
| chr13_-_44738574 | 2.29 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr21_-_36948 | 2.27 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr11_-_127785 | 2.22 |
ENSDART00000162658
|
ARFGAP1
|
ADP ribosylation factor GTPase activating protein 1 |
| chr3_+_43086548 | 2.21 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr19_+_12237945 | 2.15 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr1_+_41466011 | 2.15 |
ENSDART00000135280
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr9_+_8898835 | 2.11 |
ENSDART00000147820
|
naxd
|
NAD(P)HX dehydratase |
| chr2_-_48255843 | 2.10 |
ENSDART00000056277
|
gigyf2
|
GRB10 interacting GYF protein 2 |
| chr16_-_26676685 | 2.07 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr19_-_47832853 | 2.07 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr22_+_14836291 | 2.07 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr3_+_23742868 | 2.06 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr5_+_13326765 | 2.04 |
ENSDART00000090851
|
ydjc
|
YdjC chitooligosaccharide deacetylase homolog |
| chr7_+_52811291 | 2.04 |
ENSDART00000192900
|
CT030160.1
|
|
| chr5_+_20437124 | 2.03 |
ENSDART00000159001
|
tmem119a
|
transmembrane protein 119a |
| chr21_-_43428040 | 2.02 |
ENSDART00000148325
|
stk26
|
serine/threonine protein kinase 26 |
| chr14_-_24001825 | 2.00 |
ENSDART00000130704
|
il4
|
interleukin 4 |
| chr4_-_9557186 | 1.96 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr20_-_26537171 | 1.96 |
ENSDART00000061926
|
stx11b.2
|
syntaxin 11b, tandem duplicate 2 |
| chr3_-_40301467 | 1.95 |
ENSDART00000055186
|
atp5mf
|
ATP synthase membrane subunit f |
| chr12_-_7824291 | 1.94 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr10_+_26612321 | 1.93 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr17_+_12865746 | 1.92 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr18_+_13315739 | 1.90 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr7_+_24393678 | 1.89 |
ENSDART00000188690
|
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr13_+_28819768 | 1.88 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr10_-_35186310 | 1.88 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr8_+_36560019 | 1.87 |
ENSDART00000136418
ENSDART00000061378 ENSDART00000185237 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr7_-_59564011 | 1.82 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr14_+_22132896 | 1.79 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr12_-_26587231 | 1.78 |
ENSDART00000153168
|
slc16a5a
|
solute carrier family 16 (monocarboxylate transporter), member 5a |
| chr16_-_25680666 | 1.77 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr17_+_654759 | 1.76 |
ENSDART00000193703
|
CABZ01079748.1
|
|
| chr3_+_20012891 | 1.74 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr23_-_43486714 | 1.74 |
ENSDART00000169726
|
e2f1
|
E2F transcription factor 1 |
| chr1_-_49947290 | 1.73 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr5_+_22264051 | 1.72 |
ENSDART00000143314
|
arhgap20b
|
Rho GTPase activating protein 20b |
| chr23_+_42336084 | 1.67 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr25_+_13406069 | 1.67 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr3_+_23743139 | 1.64 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr2_+_37762924 | 1.64 |
ENSDART00000156530
|
ftr66
|
finTRIM family, member 66 |
| chr19_+_48019070 | 1.62 |
ENSDART00000158269
|
UBE2M
|
si:ch1073-205c8.3 |
| chr6_-_11812224 | 1.62 |
ENSDART00000150989
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr20_-_43742822 | 1.62 |
ENSDART00000181729
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr6_+_202367 | 1.60 |
ENSDART00000168232
|
mgat3b
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase b |
| chr1_+_10297027 | 1.60 |
ENSDART00000152562
|
eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr13_+_11876437 | 1.60 |
ENSDART00000179753
|
trim8a
|
tripartite motif containing 8a |
| chr22_-_10752471 | 1.59 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr16_+_31922065 | 1.59 |
ENSDART00000131661
ENSDART00000144194 ENSDART00000145510 |
rps9
|
ribosomal protein S9 |
| chr16_+_16529748 | 1.59 |
ENSDART00000029579
|
ccdc12
|
coiled-coil domain containing 12 |
| chr6_-_41536323 | 1.57 |
ENSDART00000113157
|
hemk1
|
HemK methyltransferase family member 1 |
| chr9_+_21277846 | 1.57 |
ENSDART00000139620
ENSDART00000110996 ENSDART00000111899 |
lats2
|
large tumor suppressor kinase 2 |
| chr23_-_4705110 | 1.55 |
ENSDART00000144536
ENSDART00000129050 ENSDART00000136399 |
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr10_-_27197044 | 1.55 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr17_+_49091661 | 1.54 |
ENSDART00000177166
ENSDART00000177390 ENSDART00000190114 |
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr1_+_8521323 | 1.54 |
ENSDART00000121439
ENSDART00000103626 ENSDART00000141283 |
mief2
|
mitochondrial elongation factor 2 |
| chr15_+_11825366 | 1.53 |
ENSDART00000190042
|
prkd2
|
protein kinase D2 |
| chr6_+_25213934 | 1.52 |
ENSDART00000163440
|
si:ch73-97h19.2
|
si:ch73-97h19.2 |
| chr12_-_7807298 | 1.52 |
ENSDART00000191355
|
ank3b
|
ankyrin 3b |
| chr20_+_30939178 | 1.51 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr3_+_54581987 | 1.51 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr6_-_40882880 | 1.50 |
ENSDART00000181706
|
sirt4
|
sirtuin 4 |
| chr24_-_2312868 | 1.46 |
ENSDART00000140125
ENSDART00000138432 |
cul2
|
cullin 2 |
| chr1_-_24918686 | 1.46 |
ENSDART00000148076
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr16_-_24550029 | 1.46 |
ENSDART00000108590
|
si:ch211-79k12.1
|
si:ch211-79k12.1 |
| chr3_+_32411343 | 1.46 |
ENSDART00000186287
ENSDART00000141793 |
rras
|
RAS related |
| chr4_-_5831036 | 1.44 |
ENSDART00000166232
|
foxm1
|
forkhead box M1 |
| chr4_+_6032640 | 1.42 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr15_-_4616816 | 1.40 |
ENSDART00000160191
ENSDART00000161721 ENSDART00000144949 |
eif4h
|
eukaryotic translation initiation factor 4h |
| chr16_+_20055878 | 1.39 |
ENSDART00000146436
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr3_+_34149337 | 1.39 |
ENSDART00000006091
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr3_+_22591395 | 1.39 |
ENSDART00000190526
|
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr12_+_36109507 | 1.39 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr18_+_44673990 | 1.38 |
ENSDART00000018625
|
napab
|
N-ethylmaleimide-sensitive factor attachment protein, alpha b |
| chr11_-_29657947 | 1.35 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr13_+_51710725 | 1.33 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr3_-_22366562 | 1.33 |
ENSDART00000129447
|
ifnphi1
|
interferon phi 1 |
| chr17_-_50022827 | 1.31 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr14_+_31493306 | 1.30 |
ENSDART00000138341
|
phf6
|
PHD finger protein 6 |
| chr16_-_46578523 | 1.30 |
ENSDART00000131061
|
si:dkey-152b24.6
|
si:dkey-152b24.6 |
| chr13_+_45524475 | 1.29 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr8_+_10869183 | 1.28 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr7_+_9981757 | 1.28 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr2_-_38080075 | 1.27 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr8_+_52377516 | 1.26 |
ENSDART00000115398
|
arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr9_+_21306902 | 1.24 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr23_-_16737161 | 1.24 |
ENSDART00000132573
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr10_-_8672820 | 1.23 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr7_-_66126628 | 1.23 |
ENSDART00000184492
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr21_+_45816030 | 1.23 |
ENSDART00000187056
|
pitx1
|
paired-like homeodomain 1 |
| chr3_-_31619463 | 1.22 |
ENSDART00000124559
|
moto
|
minamoto |
| chr5_-_13645995 | 1.22 |
ENSDART00000099665
ENSDART00000166957 |
purba
|
purine-rich element binding protein Ba |
| chr11_-_39105253 | 1.22 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr21_-_3007412 | 1.21 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr5_+_30520249 | 1.20 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr2_+_24936766 | 1.20 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr6_-_138392 | 1.18 |
ENSDART00000148974
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr5_-_16472719 | 1.17 |
ENSDART00000162071
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr18_-_21177674 | 1.16 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr18_-_24988645 | 1.16 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr18_-_5111449 | 1.14 |
ENSDART00000046902
|
pdcd10a
|
programmed cell death 10a |
| chr15_-_36347858 | 1.14 |
ENSDART00000155274
ENSDART00000157936 |
si:dkey-23k10.2
|
si:dkey-23k10.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf711
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 46.8 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 3.1 | 12.3 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 2.2 | 6.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.8 | 7.2 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 1.8 | 5.3 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 1.5 | 17.0 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 1.4 | 4.1 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 1.2 | 5.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.2 | 4.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 1.2 | 5.9 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.9 | 2.8 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.9 | 3.4 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.8 | 2.4 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.8 | 2.3 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.7 | 2.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.7 | 7.4 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.7 | 4.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.7 | 2.0 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.6 | 5.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.6 | 2.3 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.5 | 5.2 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.5 | 1.5 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.5 | 1.5 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.5 | 4.2 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.5 | 3.7 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.4 | 3.6 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.4 | 6.2 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.4 | 5.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 1.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.3 | 5.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.3 | 1.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.3 | 38.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.3 | 1.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 5.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.3 | 3.0 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 | 2.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 3.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 0.9 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.3 | 10.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 5.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.3 | 1.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 2.6 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.3 | 2.0 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.3 | 2.7 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.3 | 1.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.3 | 1.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.3 | 5.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.3 | 1.5 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.3 | 47.4 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.2 | 2.7 | GO:0030719 | P granule organization(GO:0030719) |
| 0.2 | 1.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 2.2 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.2 | 6.9 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.2 | 1.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 1.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.2 | 4.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 0.8 | GO:0061015 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.2 | 4.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 1.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 2.5 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.2 | 1.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 2.9 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.2 | 2.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.2 | 3.9 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.2 | 2.3 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.2 | 1.7 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 2.0 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.4 | GO:1990120 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 2.1 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 7.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 5.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 10.3 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.1 | 2.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.9 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.8 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 1.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 1.9 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.1 | 1.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 1.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 3.8 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 0.3 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 3.8 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.1 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 2.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.6 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.1 | 1.3 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 2.6 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.2 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.1 | 2.3 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 1.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 3.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.7 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 3.1 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.7 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 1.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 3.8 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 1.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 1.8 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.7 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 6.5 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.9 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 1.2 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.8 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 5.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 1.5 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.4 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 1.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 6.0 | GO:0010564 | regulation of cell cycle process(GO:0010564) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.6 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 1.9 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 1.5 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0032508 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) lysosomal microautophagy(GO:0016237) C-terminal protein amino acid modification(GO:0018410) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.0 | 3.4 | GO:0009953 | dorsal/ventral pattern formation(GO:0009953) |
| 0.0 | 1.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.9 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.7 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 1.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.3 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 3.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.2 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.7 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.6 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.6 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.5 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 27.4 | GO:0045095 | keratin filament(GO:0045095) |
| 1.3 | 5.2 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 1.0 | 7.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.0 | 21.4 | GO:0031430 | M band(GO:0031430) |
| 0.9 | 14.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.8 | 38.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.8 | 3.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.5 | 1.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.5 | 5.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.5 | 5.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.5 | 9.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.4 | 1.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.4 | 1.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.4 | 1.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 33.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 1.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 15.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 6.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 4.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 2.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 2.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 1.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 0.9 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 5.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 2.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 8.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 0.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 2.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 0.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 2.6 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 2.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 1.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 4.7 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 8.9 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 2.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 29.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 7.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 2.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 35.0 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 4.9 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 5.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 46.8 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 1.9 | 38.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 1.3 | 5.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.9 | 3.4 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.8 | 3.4 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.8 | 4.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.7 | 2.8 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.6 | 6.0 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.6 | 4.8 | GO:0036122 | BMP binding(GO:0036122) |
| 0.6 | 4.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 2.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.6 | 6.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 2.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.5 | 5.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 1.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.4 | 10.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.4 | 2.3 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 0.9 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.3 | 1.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.3 | 6.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 1.4 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.3 | 35.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 0.8 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.2 | 2.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 5.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 2.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.4 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.2 | 1.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.2 | 1.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 2.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.5 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 8.2 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 8.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.9 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 5.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 6.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 1.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.7 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 2.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.7 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 2.7 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 6.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 1.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 9.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 2.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 4.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 5.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 8.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 3.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 4.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 6.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 6.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 3.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 3.2 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.8 | GO:0051536 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 2.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 15.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.7 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.8 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 7.0 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.6 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 5.1 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.3 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 31.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 9.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 5.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.3 | 5.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 2.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 4.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 3.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 2.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 3.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 5.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 5.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 18.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.4 | 1.4 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.3 | 6.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.3 | 2.5 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.3 | 10.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 3.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 2.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 2.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 5.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 3.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.2 | 1.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 1.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 4.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 5.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 6.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 8.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 4.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |