Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
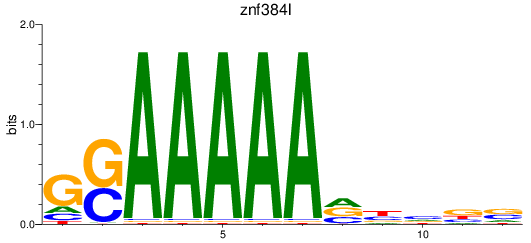
Results for znf384l
Z-value: 1.32
Transcription factors associated with znf384l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf384l
|
ENSDARG00000001015 | zinc finger protein 384 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf384l | dr11_v1_chr19_-_6983002_6983002 | -0.20 | 5.6e-02 | Click! |
Activity profile of znf384l motif
Sorted Z-values of znf384l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_1838164 | 13.46 |
ENSDART00000006013
|
atp1a1a.5
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 5 |
| chr16_-_23346095 | 12.61 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr16_+_35924188 | 11.75 |
ENSDART00000165847
|
sh3d21
|
SH3 domain containing 21 |
| chr22_-_15593824 | 10.16 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr16_-_21038015 | 10.07 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr25_+_8356707 | 9.25 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr17_-_8862424 | 9.19 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr2_-_20923864 | 8.78 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr7_-_31441420 | 8.70 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr12_-_4249000 | 8.42 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr14_-_12071679 | 8.09 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr5_+_26212621 | 7.67 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr8_+_19356072 | 7.67 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr1_-_33380011 | 7.50 |
ENSDART00000141347
ENSDART00000136383 |
cd99
|
CD99 molecule |
| chr3_-_22228602 | 7.46 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr9_-_45602978 | 7.33 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr12_+_20336070 | 7.26 |
ENSDART00000066385
|
zgc:163057
|
zgc:163057 |
| chr3_+_23221047 | 7.13 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr4_+_7677318 | 7.02 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr15_+_46356879 | 6.93 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr6_-_33075576 | 6.93 |
ENSDART00000154017
|
si:dkey-170g13.2
|
si:dkey-170g13.2 |
| chr16_-_46664465 | 6.91 |
ENSDART00000135364
|
tmem176l.4
|
transmembrane protein 176l.4 |
| chr14_+_15622817 | 6.90 |
ENSDART00000158624
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr19_-_15281996 | 6.77 |
ENSDART00000103784
|
edn2
|
endothelin 2 |
| chr19_+_7043634 | 6.67 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr11_-_42730063 | 6.54 |
ENSDART00000169776
|
si:ch73-106k19.2
|
si:ch73-106k19.2 |
| chr15_-_46779934 | 6.49 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr17_+_24851951 | 6.49 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr19_-_25519612 | 6.47 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr19_-_25005609 | 6.46 |
ENSDART00000151129
|
xkr8.2
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 2 |
| chr19_-_13808630 | 6.42 |
ENSDART00000166895
ENSDART00000187670 |
ctgfb
|
connective tissue growth factor b |
| chr14_-_12071447 | 6.28 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr3_-_39488482 | 6.27 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr22_-_17671348 | 6.26 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr1_-_59116617 | 6.18 |
ENSDART00000137471
ENSDART00000140490 |
MFAP4 (1 of many)
|
si:zfos-2330d3.7 |
| chr19_+_5640504 | 6.14 |
ENSDART00000179987
|
ft2
|
alpha(1,3)fucosyltransferase gene 2 |
| chr15_-_34892664 | 6.14 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr19_-_25519310 | 6.03 |
ENSDART00000089882
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr10_-_17988779 | 5.98 |
ENSDART00000132206
ENSDART00000144841 |
si:dkey-242g16.2
|
si:dkey-242g16.2 |
| chr7_-_38659477 | 5.77 |
ENSDART00000138071
|
npsn
|
nephrosin |
| chr16_-_12319822 | 5.71 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr2_+_36112273 | 5.62 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr19_-_977849 | 5.52 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr1_-_59096190 | 5.52 |
ENSDART00000149436
ENSDART00000191532 |
MFAP4 (1 of many)
MFAP4 (1 of many)
|
wu:fj11g02 si:zfos-2330d3.1 |
| chr20_-_35557902 | 5.50 |
ENSDART00000134753
|
adgrf7
|
adhesion G protein-coupled receptor F7 |
| chr17_+_28103675 | 5.41 |
ENSDART00000188078
|
zgc:91908
|
zgc:91908 |
| chr5_-_68876211 | 5.39 |
ENSDART00000097254
|
sftpbb
|
surfactant protein Bb |
| chr7_-_7398350 | 5.38 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr3_-_20091964 | 5.34 |
ENSDART00000029386
ENSDART00000020253 ENSDART00000124326 |
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr19_+_7045033 | 5.30 |
ENSDART00000146579
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr25_+_35502552 | 5.21 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr16_-_11932923 | 5.19 |
ENSDART00000103975
|
cd4-1
|
CD4-1 molecule |
| chr9_+_6578580 | 5.13 |
ENSDART00000061577
|
fhl2a
|
four and a half LIM domains 2a |
| chr10_+_8875195 | 5.13 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr18_+_35861930 | 5.10 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr25_+_19734038 | 5.06 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr1_+_1904419 | 5.05 |
ENSDART00000142874
|
si:ch211-132g1.4
|
si:ch211-132g1.4 |
| chr8_-_46455874 | 5.04 |
ENSDART00000146985
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr6_-_47506351 | 5.02 |
ENSDART00000128700
|
im:7151449
|
im:7151449 |
| chr6_+_23809163 | 4.98 |
ENSDART00000170402
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr17_+_25444605 | 4.95 |
ENSDART00000149804
|
clic4
|
chloride intracellular channel 4 |
| chr2_+_45191049 | 4.95 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr5_-_6567464 | 4.93 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr23_+_42813415 | 4.90 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr11_-_21404358 | 4.89 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr11_-_20096018 | 4.84 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr1_+_9994811 | 4.83 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr12_-_30548244 | 4.82 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr11_-_7261717 | 4.78 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr3_+_2971852 | 4.75 |
ENSDART00000059271
|
BX004816.1
|
|
| chr1_+_41478345 | 4.73 |
ENSDART00000134626
|
dok1a
|
docking protein 1a |
| chr4_+_7817996 | 4.72 |
ENSDART00000166809
|
si:ch1073-67j19.1
|
si:ch1073-67j19.1 |
| chr21_+_25688388 | 4.66 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr7_-_2047639 | 4.65 |
ENSDART00000173892
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr24_-_25096199 | 4.61 |
ENSDART00000185076
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr15_-_20916251 | 4.58 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr2_+_23790748 | 4.52 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr6_-_29007493 | 4.51 |
ENSDART00000065139
|
gfi1ab
|
growth factor independent 1A transcription repressor b |
| chr21_-_11646878 | 4.50 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr9_-_33081978 | 4.49 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr24_+_39990695 | 4.47 |
ENSDART00000040281
|
BX323854.1
|
|
| chr23_-_5818992 | 4.47 |
ENSDART00000148730
|
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr9_+_10692905 | 4.47 |
ENSDART00000061499
|
cxcr4b
|
chemokine (C-X-C motif), receptor 4b |
| chr19_-_5345930 | 4.45 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr2_+_36109002 | 4.45 |
ENSDART00000158978
|
traj28
|
T-cell receptor alpha joining 28 |
| chr8_-_23599096 | 4.44 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr6_-_52675630 | 4.40 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr13_-_21672131 | 4.38 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr7_-_52417060 | 4.36 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr21_+_15723069 | 4.34 |
ENSDART00000149126
ENSDART00000130628 |
p2rx4a
|
purinergic receptor P2X, ligand-gated ion channel, 4a |
| chr23_+_17865953 | 4.18 |
ENSDART00000014723
ENSDART00000140302 ENSDART00000144800 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr7_+_65673885 | 4.18 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr2_-_37744951 | 4.08 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr13_+_47810211 | 4.08 |
ENSDART00000122964
|
zmp:0000001006
|
zmp:0000001006 |
| chr8_+_21353878 | 4.07 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr16_+_33142734 | 4.05 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr5_+_58550291 | 4.02 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr16_-_39570832 | 3.99 |
ENSDART00000039832
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr20_+_34130714 | 3.97 |
ENSDART00000135915
|
pla2g4ab
|
phospholipase A2, group IVAb (cytosolic, calcium-dependent) |
| chr16_+_19732543 | 3.95 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr17_-_6451801 | 3.95 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr14_+_4151379 | 3.94 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr8_+_11687254 | 3.90 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr18_-_35459996 | 3.89 |
ENSDART00000141023
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr21_+_30794351 | 3.87 |
ENSDART00000139486
|
zgc:158225
|
zgc:158225 |
| chr7_+_35238234 | 3.86 |
ENSDART00000178732
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr16_+_38201840 | 3.84 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr3_-_4591643 | 3.82 |
ENSDART00000138144
|
ftr50
|
finTRIM family, member 50 |
| chr22_-_3344613 | 3.82 |
ENSDART00000165600
|
tbxa2r
|
thromboxane A2 receptor |
| chr15_+_24563504 | 3.82 |
ENSDART00000140658
ENSDART00000130589 ENSDART00000045549 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr7_-_20241346 | 3.73 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr21_-_11996769 | 3.73 |
ENSDART00000143537
|
zgc:64106
|
zgc:64106 |
| chr5_-_4532516 | 3.71 |
ENSDART00000192398
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr7_-_26579465 | 3.69 |
ENSDART00000173820
|
si:dkey-62k3.6
|
si:dkey-62k3.6 |
| chr7_+_2228276 | 3.68 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr2_-_43635777 | 3.65 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr10_-_43771447 | 3.65 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr12_-_44016898 | 3.64 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr3_+_3139240 | 3.63 |
ENSDART00000105014
|
si:dkey-30g5.1
|
si:dkey-30g5.1 |
| chr6_+_36821621 | 3.60 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr25_+_5035343 | 3.55 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr1_+_52392511 | 3.54 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr13_-_15994419 | 3.53 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr8_-_46509194 | 3.47 |
ENSDART00000038924
|
sult1st1
|
sulfotransferase family 1, cytosolic sulfotransferase 1 |
| chr17_+_25444323 | 3.46 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr3_+_32571929 | 3.43 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr23_-_14830627 | 3.42 |
ENSDART00000134659
|
sla2
|
Src-like-adaptor 2 |
| chr8_+_22377936 | 3.41 |
ENSDART00000181548
|
si:dkey-23c22.5
|
si:dkey-23c22.5 |
| chr16_-_30878521 | 3.41 |
ENSDART00000141403
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr3_-_2091029 | 3.40 |
ENSDART00000141464
|
si:dkey-88j15.4
|
si:dkey-88j15.4 |
| chr5_-_44286987 | 3.37 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr1_-_59102320 | 3.37 |
ENSDART00000193379
|
si:zfos-2330d3.7
|
si:zfos-2330d3.7 |
| chr9_-_14055959 | 3.33 |
ENSDART00000146675
|
fer1l6
|
fer-1-like family member 6 |
| chr20_-_25369767 | 3.32 |
ENSDART00000180278
|
itsn2a
|
intersectin 2a |
| chr19_+_7124337 | 3.31 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr4_+_38550788 | 3.30 |
ENSDART00000157412
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr18_+_45645357 | 3.27 |
ENSDART00000010256
|
eif3m
|
eukaryotic translation initiation factor 3, subunit M |
| chr12_-_44010532 | 3.27 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr12_+_30360579 | 3.23 |
ENSDART00000152900
|
si:ch211-225b10.3
|
si:ch211-225b10.3 |
| chr18_+_8833251 | 3.23 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr18_-_49020066 | 3.22 |
ENSDART00000174394
|
BX663503.3
|
|
| chr18_+_20468157 | 3.21 |
ENSDART00000100665
ENSDART00000147867 ENSDART00000060302 ENSDART00000180370 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr19_-_2861444 | 3.19 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr20_-_34292607 | 3.11 |
ENSDART00000144705
|
hmcn1
|
hemicentin 1 |
| chr8_+_8936912 | 3.10 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr19_-_24267410 | 3.09 |
ENSDART00000104083
|
s100v2
|
S100 calcium binding protein V2 |
| chr6_+_36878815 | 3.09 |
ENSDART00000192482
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr7_-_8738827 | 3.09 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr15_-_39971756 | 3.07 |
ENSDART00000063789
|
rps5
|
ribosomal protein S5 |
| chr12_-_4841018 | 3.06 |
ENSDART00000166500
|
zgc:163073
|
zgc:163073 |
| chr9_-_4075620 | 3.06 |
ENSDART00000046903
|
mylka
|
myosin, light chain kinase a |
| chr2_+_43750899 | 3.01 |
ENSDART00000135836
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr15_-_35960250 | 3.00 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr3_+_25121156 | 2.97 |
ENSDART00000025724
|
zgc:63831
|
zgc:63831 |
| chr25_+_11456696 | 2.97 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr15_+_7054754 | 2.96 |
ENSDART00000149800
|
foxl2a
|
forkhead box L2a |
| chr20_-_31067306 | 2.95 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr4_+_73606482 | 2.92 |
ENSDART00000150765
|
si:ch211-165i18.2
|
si:ch211-165i18.2 |
| chr5_+_1589458 | 2.90 |
ENSDART00000167473
|
CABZ01076669.1
|
|
| chr24_+_36636208 | 2.89 |
ENSDART00000139211
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr23_+_1276006 | 2.86 |
ENSDART00000162294
|
utrn
|
utrophin |
| chr3_+_57948377 | 2.85 |
ENSDART00000047418
|
notum1a
|
notum, palmitoleoyl-protein carboxylesterase a |
| chr16_+_28383758 | 2.84 |
ENSDART00000059038
ENSDART00000141061 |
itga8
|
integrin, alpha 8 |
| chr1_-_25177086 | 2.82 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr11_-_28050559 | 2.81 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr6_-_49215790 | 2.80 |
ENSDART00000103376
ENSDART00000132347 ENSDART00000132131 ENSDART00000143252 ENSDART00000065033 |
ngfb
|
nerve growth factor b (beta polypeptide) |
| chr8_-_32805214 | 2.80 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr19_+_19786117 | 2.79 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr5_-_34609337 | 2.78 |
ENSDART00000145792
|
hexb
|
hexosaminidase B (beta polypeptide) |
| chr10_-_15053507 | 2.76 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr14_-_1355544 | 2.73 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr19_-_43639331 | 2.73 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr2_-_58183499 | 2.72 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr6_+_48206535 | 2.71 |
ENSDART00000075172
|
cttnbp2nla
|
CTTNBP2 N-terminal like a |
| chr6_+_49052741 | 2.69 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr3_-_54669185 | 2.68 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr8_-_14126646 | 2.68 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr20_+_37393134 | 2.67 |
ENSDART00000128321
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr18_-_37355666 | 2.67 |
ENSDART00000098914
|
yap1
|
Yes-associated protein 1 |
| chr15_+_19838458 | 2.66 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr5_-_28968964 | 2.65 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr1_+_20635190 | 2.65 |
ENSDART00000145418
ENSDART00000148518 ENSDART00000139461 ENSDART00000102969 ENSDART00000166479 |
spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr13_-_42673978 | 2.64 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr13_+_22264914 | 2.63 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr16_-_30903930 | 2.59 |
ENSDART00000143996
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr21_-_20832482 | 2.59 |
ENSDART00000191928
|
c6
|
complement component 6 |
| chr23_+_17865554 | 2.58 |
ENSDART00000181009
ENSDART00000162822 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr2_+_38161318 | 2.55 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr17_+_20605013 | 2.55 |
ENSDART00000156878
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr12_-_44307963 | 2.54 |
ENSDART00000161009
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr18_+_18455029 | 2.53 |
ENSDART00000165079
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr23_+_38251864 | 2.52 |
ENSDART00000183498
ENSDART00000129593 |
znf217
|
zinc finger protein 217 |
| chr2_-_45191319 | 2.50 |
ENSDART00000192272
|
CR407590.2
|
|
| chr9_-_33328948 | 2.50 |
ENSDART00000006948
|
rpl8
|
ribosomal protein L8 |
| chr16_-_32013913 | 2.50 |
ENSDART00000030282
ENSDART00000138701 |
gstk1
|
glutathione S-transferase kappa 1 |
| chr16_-_32208728 | 2.49 |
ENSDART00000023995
|
calhm5.2
|
calcium homeostasis modulator family member 5, tandem duplicate 2 |
| chr24_+_211217 | 2.49 |
ENSDART00000151925
ENSDART00000161636 |
si:dkey-225n22.4
|
si:dkey-225n22.4 |
| chr21_+_5800306 | 2.48 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr1_+_58282449 | 2.46 |
ENSDART00000131475
|
si:dkey-222h21.7
|
si:dkey-222h21.7 |
| chr3_+_24634481 | 2.45 |
ENSDART00000163080
|
si:dkey-68o6.8
|
si:dkey-68o6.8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf384l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.8 | 7.1 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 1.7 | 6.8 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.5 | 7.5 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 1.4 | 14.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 1.3 | 5.4 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 1.2 | 4.7 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 1.1 | 4.5 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 1.1 | 4.4 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 1.1 | 4.4 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 1.0 | 3.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 1.0 | 3.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.9 | 18.0 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.9 | 5.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.9 | 2.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.9 | 2.7 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.8 | 3.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.8 | 2.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.7 | 2.9 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.7 | 7.0 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.7 | 2.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.7 | 3.9 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.6 | 4.5 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.6 | 6.5 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.6 | 1.1 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.6 | 1.1 | GO:0048901 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.5 | 2.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.5 | 2.2 | GO:1990544 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.5 | 1.6 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.5 | 2.7 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.5 | 2.0 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.5 | 1.9 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.5 | 4.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.4 | 1.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.4 | 1.8 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.4 | 6.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.4 | 6.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 2.7 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.4 | 0.8 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.4 | 4.3 | GO:0033198 | response to ATP(GO:0033198) |
| 0.4 | 2.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.4 | 1.8 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.4 | 10.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 5.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 7.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 2.1 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 3.2 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.3 | 1.0 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.3 | 3.8 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.3 | 1.3 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.3 | 2.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.3 | 4.7 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.3 | 1.2 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.3 | 5.5 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.3 | 1.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.3 | 3.4 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.3 | 2.6 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 | 6.0 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.3 | 3.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 1.6 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.3 | 5.2 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.3 | 1.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.3 | 1.8 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 4.1 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 | 7.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.3 | 2.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 1.2 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.2 | 1.2 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.2 | 2.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 9.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 2.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 1.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 3.8 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 4.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 3.1 | GO:0090481 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 1.6 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 3.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 1.7 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.2 | 2.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 3.5 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.2 | 0.6 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.2 | 4.6 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.2 | 2.1 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 0.5 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.2 | 1.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 1.7 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 1.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 4.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 0.8 | GO:0071320 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 7.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.2 | 3.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.2 | 0.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 3.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 14.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 1.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 2.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 20.8 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.7 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 2.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 2.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 3.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 5.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.2 | GO:0034694 | response to prostaglandin(GO:0034694) cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 10.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 0.3 | GO:0002698 | negative regulation of immune effector process(GO:0002698) |
| 0.1 | 3.1 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.1 | 1.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 5.8 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.1 | 2.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.2 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 3.2 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 1.3 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 3.6 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 3.2 | GO:0009411 | response to UV(GO:0009411) |
| 0.1 | 1.6 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 4.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 2.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 10.0 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 0.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 3.3 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.0 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.3 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 1.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 3.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 2.2 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 9.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.9 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 0.7 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 2.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.0 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.1 | 7.5 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 1.9 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.1 | 1.2 | GO:0010810 | regulation of cell-substrate adhesion(GO:0010810) positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.3 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 2.9 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 2.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 1.7 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.8 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 1.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 0.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.5 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 1.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.7 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 1.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.1 | 2.2 | GO:0060840 | artery development(GO:0060840) |
| 0.1 | 2.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 5.0 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.1 | 5.0 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.1 | 1.9 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 2.2 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.3 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 5.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 1.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.8 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.1 | 1.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 0.3 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.1 | 0.9 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.9 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 4.6 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 2.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 1.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 2.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.6 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 2.6 | GO:0070997 | neuron death(GO:0070997) |
| 0.0 | 0.2 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 1.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.0 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.4 | GO:0009204 | nucleoside triphosphate catabolic process(GO:0009143) deoxyribonucleoside triphosphate catabolic process(GO:0009204) |
| 0.0 | 0.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 1.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.6 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.2 | GO:0032979 | respiratory chain complex III assembly(GO:0017062) protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) mitochondrial respiratory chain complex III assembly(GO:0034551) protein insertion into mitochondrial membrane(GO:0051204) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.7 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 13.4 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.6 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0085029 | extracellular matrix assembly(GO:0085029) |
| 0.0 | 3.6 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 3.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 2.4 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 1.7 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 3.3 | GO:0072594 | establishment of protein localization to organelle(GO:0072594) |
| 0.0 | 1.3 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 1.0 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.9 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 0.1 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 1.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.5 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.3 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.3 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 1.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.4 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0022406 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 1.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.3 | GO:0045597 | positive regulation of cell differentiation(GO:0045597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.8 | 3.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.7 | 3.0 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.6 | 7.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 8.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.4 | 6.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 4.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 2.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 2.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 2.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 5.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 2.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 4.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 1.0 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 9.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 2.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 3.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 4.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 1.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 0.9 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 2.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 1.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 15.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 16.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 3.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 8.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 15.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 3.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 32.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 6.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 2.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 10.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.3 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 6.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 3.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 7.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 4.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 5.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 3.1 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 10.1 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 4.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 13.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 2.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 3.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0098844 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 4.2 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0097526 | spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 4.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 28.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 3.4 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 4.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.5 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.0 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 2.2 | 6.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.7 | 6.8 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.6 | 15.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.4 | 4.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.3 | 3.9 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.9 | 8.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.9 | 4.4 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.7 | 2.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.7 | 3.7 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.7 | 14.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 2.9 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.7 | 4.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.6 | 4.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 7.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.6 | 2.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 2.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.5 | 3.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 1.6 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.5 | 6.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.5 | 3.0 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.4 | 5.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.6 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.4 | 1.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 6.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 3.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 1.9 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.4 | 2.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 4.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.4 | 4.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 4.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 1.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.3 | 2.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 2.7 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 2.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.3 | 1.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 1.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 0.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.9 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.3 | 1.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 2.6 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 3.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 0.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 3.9 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.2 | 5.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 4.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 3.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 2.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.2 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 4.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 2.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 3.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 1.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 5.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 10.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 0.9 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.6 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 0.5 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.2 | 1.5 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 4.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 1.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 6.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 3.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 0.8 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 5.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.2 | 3.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 1.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 2.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 4.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 2.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 5.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 4.5 | GO:0019956 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.1 | 1.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 2.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 8.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 3.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 1.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 2.1 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 4.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 5.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.3 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 2.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.1 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 5.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 1.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 8.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 2.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 10.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 9.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 5.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.9 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 2.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 10.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 2.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 11.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0005227 | calcium activated cation channel activity(GO:0005227) ion gated channel activity(GO:0022839) |
| 0.0 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 2.8 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 11.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.6 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 2.6 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 9.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 3.2 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 5.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 3.4 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 1.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 7.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 2.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 3.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 4.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 2.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 3.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 1.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 0.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 2.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 3.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 2.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 4.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 9.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 1.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 5.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 5.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 4.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 11.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 2.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.5 | 2.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 4.9 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 5.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 2.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 3.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 4.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 4.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 4.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 1.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 7.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 6.7 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.1 | 2.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 3.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.1 | 2.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 3.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 5.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 6.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 9.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.8 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 3.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.8 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 3.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.1 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 2.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 2.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 3.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |