Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
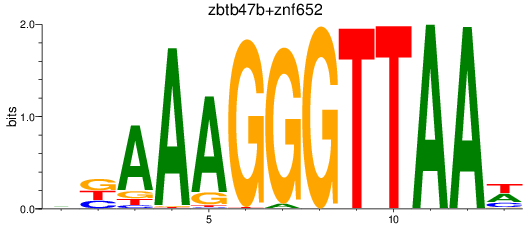
Results for zbtb47b+znf652
Z-value: 1.68
Transcription factors associated with zbtb47b+znf652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf652
|
ENSDARG00000062302 | zinc finger protein 652 |
|
zbtb47b
|
ENSDARG00000079547 | zinc finger and BTB domain containing 47b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb47b | dr11_v1_chr24_-_20599781_20599880 | -0.59 | 3.7e-10 | Click! |
| znf652 | dr11_v1_chr3_+_15773991_15773991 | -0.57 | 2.9e-09 | Click! |
Activity profile of zbtb47b+znf652 motif
Sorted Z-values of zbtb47b+znf652 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23913943 | 46.68 |
ENSDART00000175404
ENSDART00000129525 |
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr21_+_27416284 | 33.26 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr16_-_31661536 | 32.29 |
ENSDART00000169973
|
wu:fd46c06
|
wu:fd46c06 |
| chr13_-_18637244 | 26.08 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr22_-_36856405 | 24.65 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr5_+_37978501 | 23.85 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr16_-_31675669 | 23.10 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr3_-_54544612 | 20.78 |
ENSDART00000018044
|
angptl6
|
angiopoietin-like 6 |
| chr16_-_22294265 | 20.65 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr5_-_69948099 | 20.09 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr8_-_30791089 | 18.83 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr8_-_30791266 | 17.86 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr11_+_13223625 | 16.91 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr14_+_32430982 | 16.29 |
ENSDART00000017179
ENSDART00000123382 ENSDART00000075593 |
f9a
|
coagulation factor IXa |
| chr14_-_83154 | 16.09 |
ENSDART00000187097
|
CABZ01086812.1
|
|
| chr8_+_1766206 | 16.03 |
ENSDART00000021820
|
serpind1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr21_+_20771082 | 15.95 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr7_-_26306546 | 14.26 |
ENSDART00000140817
|
zgc:77439
|
zgc:77439 |
| chr7_-_26306954 | 13.65 |
ENSDART00000057288
|
zgc:77439
|
zgc:77439 |
| chr23_+_42454292 | 12.99 |
ENSDART00000171459
|
cyp2aa2
|
y cytochrome P450, family 2, subfamily AA, polypeptide 2 |
| chr25_+_36674715 | 12.43 |
ENSDART00000111861
|
mafb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
| chr7_+_58699900 | 12.36 |
ENSDART00000144009
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr21_-_27443995 | 11.80 |
ENSDART00000003508
|
bfb
|
complement component bfb |
| chr7_+_58699718 | 11.34 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr21_-_40083432 | 10.69 |
ENSDART00000141160
ENSDART00000191195 |
slc13a5a
|
info solute carrier family 13 (sodium-dependent citrate transporter), member 5a |
| chr3_-_39198113 | 10.48 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr14_-_46897067 | 10.22 |
ENSDART00000058789
|
qdpra
|
quinoid dihydropteridine reductase a |
| chr20_-_39219537 | 9.05 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr20_-_19365875 | 8.83 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr18_-_977075 | 8.55 |
ENSDART00000032392
|
dhdhl
|
dihydrodiol dehydrogenase (dimeric), like |
| chr23_+_26026383 | 8.00 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr10_+_4987766 | 7.90 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr24_-_31332087 | 7.23 |
ENSDART00000161179
|
abcd3a
|
ATP-binding cassette, sub-family D (ALD), member 3a |
| chr7_+_54222156 | 7.00 |
ENSDART00000165201
ENSDART00000158518 |
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr5_-_42904329 | 6.91 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr18_-_14836862 | 6.62 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr12_-_48671612 | 6.21 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr8_-_16609004 | 6.05 |
ENSDART00000102556
|
tor3a
|
torsin family 3, member A |
| chr13_+_844150 | 5.99 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr7_-_7823662 | 5.94 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr10_+_11260170 | 5.82 |
ENSDART00000155742
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr6_-_2627488 | 5.77 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr11_-_45138857 | 5.72 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr19_-_27391179 | 5.70 |
ENSDART00000181108
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr5_+_4298636 | 5.69 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr13_+_835390 | 5.61 |
ENSDART00000171329
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr3_+_49043917 | 5.52 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr20_+_51479263 | 5.42 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr8_-_32385989 | 5.42 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr9_+_38408991 | 5.32 |
ENSDART00000045588
|
cyp27a7
|
cytochrome P450, family 27, subfamily A, polypeptide 7 |
| chr9_+_38420028 | 5.27 |
ENSDART00000135748
|
cyp27a1.2
|
cytochrome P450, family 27, subfamily A, polypeptide 1, gene 2 |
| chr11_-_23458792 | 5.22 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr22_+_20135443 | 5.18 |
ENSDART00000143641
|
eef2a.1
|
eukaryotic translation elongation factor 2a, tandem duplicate 1 |
| chr13_+_7442023 | 5.17 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr23_+_42434348 | 5.12 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr7_+_56615554 | 5.02 |
ENSDART00000098430
|
dpep1
|
dipeptidase 1 |
| chr23_+_7692042 | 4.99 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr19_-_23249822 | 4.89 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr1_-_7951002 | 4.87 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr21_+_17768174 | 4.70 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr12_-_31461412 | 4.65 |
ENSDART00000175929
ENSDART00000186342 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr7_+_36539124 | 4.57 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr6_+_34028532 | 4.46 |
ENSDART00000155827
|
si:ch73-185c24.2
|
si:ch73-185c24.2 |
| chr9_+_38427572 | 4.40 |
ENSDART00000108860
|
CYP27A1 (1 of many)
|
zgc:136333 |
| chr12_-_4220713 | 4.36 |
ENSDART00000129427
|
vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr11_+_3959495 | 4.36 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr12_-_31461219 | 4.35 |
ENSDART00000148954
|
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr16_-_12236362 | 4.34 |
ENSDART00000114759
|
lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr7_+_61764040 | 4.33 |
ENSDART00000056745
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr16_+_35595312 | 4.27 |
ENSDART00000170438
|
si:ch211-1i11.3
|
si:ch211-1i11.3 |
| chr22_+_5682635 | 4.22 |
ENSDART00000140680
ENSDART00000131308 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr2_+_52232630 | 4.21 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr17_-_53353653 | 4.19 |
ENSDART00000180744
ENSDART00000026879 |
unm_sa911
|
un-named sa911 |
| chr7_+_19482084 | 4.17 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr8_-_38810233 | 4.13 |
ENSDART00000085304
|
pcsk5b
|
proprotein convertase subtilisin/kexin type 5b |
| chr11_+_38429319 | 4.08 |
ENSDART00000029157
|
mfsd4aa
|
major facilitator superfamily domain containing 4Aa |
| chr24_-_27423697 | 3.92 |
ENSDART00000132590
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr12_+_2648043 | 3.92 |
ENSDART00000082220
|
gdf2
|
growth differentiation factor 2 |
| chr7_+_17947217 | 3.92 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr23_-_17101360 | 3.91 |
ENSDART00000053418
|
dnmt3bb.1
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.1 |
| chr17_-_25737452 | 3.89 |
ENSDART00000152021
|
si:ch211-214p16.3
|
si:ch211-214p16.3 |
| chr7_+_37377335 | 3.89 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr21_-_22435957 | 3.88 |
ENSDART00000137959
|
il7r
|
interleukin 7 receptor |
| chr9_+_38446504 | 3.87 |
ENSDART00000077479
|
cyp27a1.4
|
cytochrome P450, family 27, subfamily A, polypeptide 1, gene 4 |
| chr1_+_41478345 | 3.85 |
ENSDART00000134626
|
dok1a
|
docking protein 1a |
| chr17_+_6536152 | 3.84 |
ENSDART00000062952
ENSDART00000121789 |
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr20_+_51478939 | 3.78 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr10_+_9550419 | 3.76 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr16_+_3982590 | 3.71 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr3_+_18097700 | 3.68 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr3_-_54846444 | 3.62 |
ENSDART00000074010
|
ubald1b
|
UBA-like domain containing 1b |
| chr22_-_36774219 | 3.60 |
ENSDART00000056151
ENSDART00000168711 |
acy1
|
aminoacylase 1 |
| chr2_+_38271392 | 3.56 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr10_+_2600830 | 3.56 |
ENSDART00000101012
|
CU856539.2
|
|
| chr4_+_58576146 | 3.49 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr16_-_38609146 | 3.47 |
ENSDART00000144651
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr17_+_5351922 | 3.45 |
ENSDART00000105394
|
runx2a
|
runt-related transcription factor 2a |
| chr4_-_71554068 | 3.45 |
ENSDART00000167414
|
si:dkey-27n6.1
|
si:dkey-27n6.1 |
| chr6_-_43677125 | 3.42 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr10_-_21545091 | 3.34 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr4_-_44673017 | 3.23 |
ENSDART00000156670
|
si:dkey-7j22.4
|
si:dkey-7j22.4 |
| chr16_+_19013018 | 3.15 |
ENSDART00000191526
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr4_+_33461796 | 3.14 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr15_+_5299404 | 3.11 |
ENSDART00000155410
|
or122-2
|
odorant receptor, family E, subfamily 122, member 2 |
| chr4_+_42175261 | 3.04 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr10_+_40660772 | 2.98 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr4_+_62431132 | 2.97 |
ENSDART00000160226
|
si:ch211-79g12.1
|
si:ch211-79g12.1 |
| chr16_-_29557338 | 2.88 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr3_-_27880229 | 2.87 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr20_-_47270519 | 2.86 |
ENSDART00000153329
|
si:dkeyp-104f11.8
|
si:dkeyp-104f11.8 |
| chr3_+_49521106 | 2.85 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr11_+_2398843 | 2.83 |
ENSDART00000126761
|
pip4k2cb
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma b |
| chr10_+_8534750 | 2.82 |
ENSDART00000183960
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr20_-_4793450 | 2.82 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr3_+_21225750 | 2.79 |
ENSDART00000139213
|
zgc:153968
|
zgc:153968 |
| chr6_+_9427641 | 2.77 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr4_+_64339299 | 2.76 |
ENSDART00000191590
|
si:ch211-223a21.1
|
si:ch211-223a21.1 |
| chr16_-_42056137 | 2.72 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr4_+_40035564 | 2.72 |
ENSDART00000168982
ENSDART00000172398 ENSDART00000163041 |
si:dkey-238g7.1
|
si:dkey-238g7.1 |
| chr22_-_24946883 | 2.71 |
ENSDART00000193725
ENSDART00000187812 |
si:dkey-4c23.5
|
si:dkey-4c23.5 |
| chr4_-_49133107 | 2.69 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr7_-_45993416 | 2.67 |
ENSDART00000157602
ENSDART00000166983 |
zgc:101715
|
zgc:101715 |
| chr17_+_26815021 | 2.60 |
ENSDART00000086885
|
asmt2
|
acetylserotonin O-methyltransferase 2 |
| chr15_-_28805493 | 2.57 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr5_+_36932718 | 2.56 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr18_+_16426266 | 2.56 |
ENSDART00000136626
|
si:dkey-287h13.1
|
si:dkey-287h13.1 |
| chr2_-_13254594 | 2.53 |
ENSDART00000155671
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr13_+_13930263 | 2.53 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr4_+_29344957 | 2.52 |
ENSDART00000185164
|
CR847895.1
|
|
| chr17_-_6600899 | 2.52 |
ENSDART00000154074
ENSDART00000180912 |
ANKRD66
|
si:ch211-189e2.2 |
| chr5_+_58455488 | 2.50 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr15_-_36357889 | 2.50 |
ENSDART00000156377
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr6_-_38816500 | 2.43 |
ENSDART00000190866
ENSDART00000104124 |
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr6_+_31368071 | 2.42 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr17_-_45125537 | 2.40 |
ENSDART00000113552
|
zgc:163014
|
zgc:163014 |
| chr3_-_43959873 | 2.40 |
ENSDART00000161582
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr4_-_29833326 | 2.37 |
ENSDART00000150596
|
si:ch211-231i17.4
|
si:ch211-231i17.4 |
| chr4_+_33462238 | 2.37 |
ENSDART00000111083
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr17_+_5426087 | 2.36 |
ENSDART00000131430
ENSDART00000188636 |
runx2a
|
runt-related transcription factor 2a |
| chr4_+_45504471 | 2.34 |
ENSDART00000150399
|
si:dkey-256i11.2
|
si:dkey-256i11.2 |
| chr4_+_64147241 | 2.32 |
ENSDART00000163509
|
znf1089
|
zinc finger protein 1089 |
| chr10_+_40646764 | 2.31 |
ENSDART00000184280
ENSDART00000135939 |
taar19s
|
trace amine associated receptor 19s |
| chr15_-_8856391 | 2.29 |
ENSDART00000008273
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr19_+_7575341 | 2.29 |
ENSDART00000134271
|
s100u
|
S100 calcium binding protein U |
| chr4_+_59711338 | 2.28 |
ENSDART00000150849
|
si:dkey-149m13.4
|
si:dkey-149m13.4 |
| chr2_-_55853943 | 2.28 |
ENSDART00000122576
|
rx2
|
retinal homeobox gene 2 |
| chr16_+_45425181 | 2.27 |
ENSDART00000168591
|
phf1
|
PHD finger protein 1 |
| chr8_+_8643901 | 2.27 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr11_-_45141309 | 2.23 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr1_+_31573225 | 2.23 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr4_+_35901451 | 2.21 |
ENSDART00000131142
|
znf1088
|
zinc finger protein 1088 |
| chr1_-_49505449 | 2.21 |
ENSDART00000187294
ENSDART00000132171 |
si:dkeyp-80c12.5
|
si:dkeyp-80c12.5 |
| chr4_+_34657691 | 2.21 |
ENSDART00000158655
|
AL645691.1
|
|
| chr5_+_45918680 | 2.21 |
ENSDART00000036242
|
ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr8_-_39822917 | 2.20 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr17_-_15747296 | 2.19 |
ENSDART00000153970
|
cx52.7
|
connexin 52.7 |
| chr4_+_40858450 | 2.19 |
ENSDART00000152047
|
si:dkey-285e18.6
|
si:dkey-285e18.6 |
| chr21_-_2814709 | 2.19 |
ENSDART00000097664
|
SEMA4D
|
semaphorin 4D |
| chr22_-_24946694 | 2.18 |
ENSDART00000137323
|
si:dkey-4c23.5
|
si:dkey-4c23.5 |
| chr20_+_27646772 | 2.18 |
ENSDART00000141697
|
mthfd1a
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1a, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr4_-_40536509 | 2.17 |
ENSDART00000150835
|
si:dkey-57c22.1
|
si:dkey-57c22.1 |
| chr4_-_50930911 | 2.17 |
ENSDART00000193337
|
si:ch211-208f21.3
|
si:ch211-208f21.3 |
| chr4_+_41082518 | 2.17 |
ENSDART00000151846
|
si:dkey-16p19.1
|
si:dkey-16p19.1 |
| chr4_-_26095755 | 2.17 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr14_+_41576849 | 2.17 |
ENSDART00000074424
|
nitr13
|
novel immune-type receptor 13 |
| chr4_+_60547709 | 2.16 |
ENSDART00000158732
|
si:dkey-211i20.5
|
si:dkey-211i20.5 |
| chr7_-_6427198 | 2.15 |
ENSDART00000173446
|
FP340257.1
|
|
| chr12_-_33770299 | 2.14 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr17_-_29194219 | 2.14 |
ENSDART00000157340
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr4_-_45433060 | 2.14 |
ENSDART00000150524
|
si:ch211-162i8.4
|
si:ch211-162i8.4 |
| chr1_-_46505310 | 2.12 |
ENSDART00000178072
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr2_-_13254821 | 2.11 |
ENSDART00000022621
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr4_-_16345227 | 2.09 |
ENSDART00000079521
|
kera
|
keratocan |
| chr1_+_49955869 | 2.09 |
ENSDART00000150517
|
gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr12_+_31775182 | 2.07 |
ENSDART00000192235
|
lrrc59
|
leucine rich repeat containing 59 |
| chr19_+_24575077 | 2.05 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr22_+_26263290 | 2.05 |
ENSDART00000184840
|
BX649315.1
|
|
| chr4_-_45301719 | 2.05 |
ENSDART00000150282
|
si:ch211-162i8.2
|
si:ch211-162i8.2 |
| chr24_-_40860603 | 2.03 |
ENSDART00000188032
|
CU633479.7
|
|
| chr4_-_42294516 | 2.03 |
ENSDART00000133558
|
si:dkey-4e4.1
|
si:dkey-4e4.1 |
| chr10_+_29849977 | 2.01 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr4_+_69191065 | 2.01 |
ENSDART00000170595
|
znf1075
|
zinc finger protein 1075 |
| chr4_+_41082985 | 2.01 |
ENSDART00000164388
|
si:dkey-16p19.1
|
si:dkey-16p19.1 |
| chr6_+_21684296 | 2.00 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr4_-_63769147 | 1.99 |
ENSDART00000189919
|
znf1098
|
zinc finger protein 1098 |
| chr5_+_27897504 | 1.98 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr4_+_63818718 | 1.97 |
ENSDART00000161177
|
si:dkey-30f3.2
|
si:dkey-30f3.2 |
| chr25_+_19680962 | 1.97 |
ENSDART00000167260
|
si:dkeyp-110c12.3
|
si:dkeyp-110c12.3 |
| chr1_-_29658721 | 1.97 |
ENSDART00000132063
|
si:dkey-1h24.6
|
si:dkey-1h24.6 |
| chr8_+_52377516 | 1.97 |
ENSDART00000115398
|
arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr7_-_19544575 | 1.97 |
ENSDART00000161549
|
CCNB1IP1
|
si:ch211-212k18.11 |
| chr4_+_59748607 | 1.96 |
ENSDART00000108499
|
znf1068
|
zinc finger protein 1068 |
| chr16_+_33902006 | 1.95 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr22_+_18349794 | 1.95 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr4_-_43280244 | 1.95 |
ENSDART00000150762
|
si:dkeyp-53e4.1
|
si:dkeyp-53e4.1 |
| chr20_-_13765749 | 1.94 |
ENSDART00000133529
|
opn8c
|
opsin 8, group member c |
| chr4_-_30528239 | 1.93 |
ENSDART00000169290
|
znf1052
|
zinc finger protein 1052 |
| chr20_+_25625872 | 1.93 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr2_-_37312927 | 1.92 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr4_-_57416684 | 1.92 |
ENSDART00000169434
|
si:dkey-122c11.7
|
si:dkey-122c11.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb47b+znf652
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.5 | GO:0042368 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 4.8 | 23.9 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 4.2 | 16.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 3.7 | 26.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 3.5 | 24.7 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 3.2 | 15.9 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 3.0 | 9.0 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 2.3 | 9.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 1.5 | 9.0 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 1.5 | 4.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 1.3 | 7.9 | GO:0070254 | mucus secretion(GO:0070254) |
| 1.3 | 11.8 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 1.2 | 7.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 1.2 | 4.6 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 1.2 | 48.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 1.1 | 4.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.0 | 9.9 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 1.0 | 3.8 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 1.0 | 2.9 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.9 | 3.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.9 | 2.6 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.8 | 2.5 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.8 | 5.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.8 | 5.8 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.8 | 12.9 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.8 | 3.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.7 | 7.0 | GO:0014028 | notochord formation(GO:0014028) |
| 0.6 | 3.9 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.6 | 2.9 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.5 | 4.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.5 | 5.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.5 | 2.5 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.5 | 8.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.5 | 1.5 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.5 | 2.0 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.5 | 46.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.5 | 32.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.5 | 1.4 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.4 | 4.4 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.4 | 2.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.4 | 42.9 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.4 | 23.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.4 | 3.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.4 | 2.2 | GO:0097065 | anterior head development(GO:0097065) |
| 0.4 | 4.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.4 | 2.8 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.3 | 6.3 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.3 | 3.9 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 | 5.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 6.2 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 45.2 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.3 | 1.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 0.9 | GO:0071706 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.3 | 1.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.3 | 4.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.3 | 5.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 4.2 | GO:0006308 | DNA catabolic process, endonucleolytic(GO:0000737) DNA catabolic process(GO:0006308) |
| 0.2 | 4.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.2 | 1.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.0 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 4.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 2.7 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.7 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 2.3 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.1 | 2.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 3.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 1.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 1.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 3.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 3.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 5.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.9 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.3 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 2.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.8 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 1.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 7.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 0.2 | GO:0045933 | positive regulation of muscle contraction(GO:0045933) positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 6.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 1.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 1.9 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 2.1 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 3.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.7 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 1.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.9 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 2.6 | GO:2000027 | regulation of organ morphogenesis(GO:2000027) |
| 0.0 | 2.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 2.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.4 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.3 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 8.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.4 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 2.8 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 3.4 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 1.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.2 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 1.8 | 23.9 | GO:0042627 | chylomicron(GO:0042627) |
| 0.8 | 3.0 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.7 | 3.5 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.5 | 1.4 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 23.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.4 | 1.7 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.3 | 2.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 1.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 1.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 7.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 10.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 6.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 2.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 8.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 5.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 161.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 4.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 8.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 51.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.3 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 26.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 5.2 | 20.6 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 4.8 | 14.5 | GO:0031073 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 4.8 | 23.9 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 4.2 | 16.9 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 3.2 | 12.9 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 3.2 | 15.9 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 2.7 | 24.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 2.6 | 10.2 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 2.5 | 9.9 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 1.4 | 48.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 1.4 | 5.7 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 1.0 | 5.0 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.0 | 3.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.9 | 4.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.9 | 4.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.8 | 2.5 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.8 | 9.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.8 | 5.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 24.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.6 | 8.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.6 | 8.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.5 | 4.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 1.5 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.5 | 5.4 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.5 | 7.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 1.4 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.4 | 5.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 4.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.4 | 4.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.4 | 93.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.4 | 21.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.4 | 1.5 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.3 | 3.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 3.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 1.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 24.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 2.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.3 | 8.0 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.3 | 3.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.3 | 6.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 1.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 5.0 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.3 | 3.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 1.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.2 | 0.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 1.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 32.2 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.2 | 2.4 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 0.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.2 | 4.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 1.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 1.8 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 2.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 11.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 17.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 4.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 5.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.2 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 2.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 2.3 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 3.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 2.8 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 1.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 2.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 6.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 4.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 3.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 4.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 0.9 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 35.9 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 1.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 3.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.9 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 58.7 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 2.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.0 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 2.8 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 2.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 33.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 1.9 | 24.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.5 | 2.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 3.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 3.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 1.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 19.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 19.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 33.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.8 | 9.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.6 | 24.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 1.1 | 9.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 1.0 | 26.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.9 | 4.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.7 | 4.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.6 | 6.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.5 | 6.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 8.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 1.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 4.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 5.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.9 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 2.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |