Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
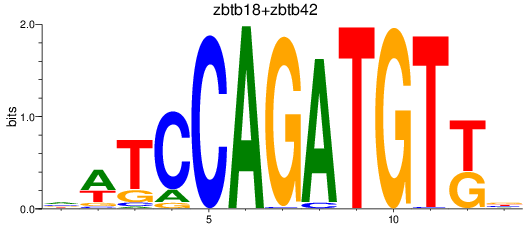
Results for zbtb18+zbtb42
Z-value: 1.20
Transcription factors associated with zbtb18+zbtb42
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb18
|
ENSDARG00000028228 | zinc finger and BTB domain containing 18 |
|
zbtb42
|
ENSDARG00000102761 | zinc finger and BTB domain containing 42 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb18 | dr11_v1_chr13_+_11436130_11436130 | 0.15 | 1.4e-01 | Click! |
| zbtb42 | dr11_v1_chr17_-_1407593_1407725 | 0.01 | 8.9e-01 | Click! |
Activity profile of zbtb18+zbtb42 motif
Sorted Z-values of zbtb18+zbtb42 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_27857051 | 11.58 |
ENSDART00000043462
ENSDART00000137861 |
acvrl1
|
activin A receptor like type 1 |
| chr15_-_33834577 | 10.54 |
ENSDART00000163354
|
mmp13b
|
matrix metallopeptidase 13b |
| chr15_-_34567370 | 10.19 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr10_-_15128771 | 9.98 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr17_+_42274825 | 9.91 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr18_-_6982499 | 9.54 |
ENSDART00000101525
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr1_+_40724140 | 9.44 |
ENSDART00000074839
|
CU550714.1
|
|
| chr25_+_35502552 | 8.68 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr17_+_42274240 | 8.58 |
ENSDART00000134377
|
pax1a
|
paired box 1a |
| chr10_+_8968203 | 7.75 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr16_-_26676685 | 7.74 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr19_-_35596207 | 7.43 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr11_-_24681292 | 6.55 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr3_-_32596394 | 6.26 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr4_+_4232562 | 6.15 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr5_+_30741730 | 5.74 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr9_-_9842149 | 5.65 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr6_+_23810529 | 5.63 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr22_-_28653074 | 5.55 |
ENSDART00000154717
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr1_+_45351890 | 5.53 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr5_+_32924669 | 5.52 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr24_-_25166720 | 5.47 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr3_-_54669185 | 5.24 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr20_+_16750177 | 5.21 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr4_+_15944245 | 5.07 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr16_-_13595027 | 5.05 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr18_+_48423973 | 4.97 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr9_-_38587275 | 4.84 |
ENSDART00000077446
|
si:dkey-101k6.5
|
si:dkey-101k6.5 |
| chr20_-_20932760 | 4.82 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr6_-_29195642 | 4.74 |
ENSDART00000078625
|
dpt
|
dermatopontin |
| chr1_+_58303892 | 4.69 |
ENSDART00000147678
|
CR769768.1
|
|
| chr20_+_23238833 | 4.69 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr1_-_11973341 | 4.64 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr13_-_13294847 | 4.49 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr22_-_26558166 | 4.37 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr25_+_753364 | 4.35 |
ENSDART00000183804
|
TWF1 (1 of many)
|
twinfilin actin binding protein 1 |
| chr3_-_36839115 | 4.25 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr19_+_24882845 | 4.13 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr16_+_9609721 | 4.12 |
ENSDART00000047920
|
enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_-_39474235 | 4.05 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr11_+_10548171 | 4.04 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr23_-_452365 | 3.92 |
ENSDART00000146776
|
tspan2b
|
tetraspanin 2b |
| chr15_-_25571865 | 3.88 |
ENSDART00000077836
|
mmp20b
|
matrix metallopeptidase 20b (enamelysin) |
| chr10_-_22803740 | 3.82 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr13_+_12045475 | 3.81 |
ENSDART00000163053
ENSDART00000160812 ENSDART00000158244 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr12_-_28881638 | 3.77 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr19_+_1688727 | 3.71 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr8_-_9570511 | 3.71 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr7_+_56889879 | 3.71 |
ENSDART00000039810
|
myo9aa
|
myosin IXAa |
| chr19_+_47502581 | 3.65 |
ENSDART00000171526
|
CU695215.2
|
|
| chr20_-_13673566 | 3.64 |
ENSDART00000181641
|
TAGAP
|
si:ch211-122h15.4 |
| chr21_+_22878834 | 3.61 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr9_+_22764235 | 3.54 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr11_+_12719944 | 3.39 |
ENSDART00000054837
ENSDART00000144954 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr16_-_50952266 | 3.35 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr18_-_46241775 | 3.32 |
ENSDART00000145999
ENSDART00000134244 ENSDART00000185021 ENSDART00000181855 |
prx
|
periaxin |
| chr19_-_24267823 | 3.31 |
ENSDART00000132430
|
s100v2
|
S100 calcium binding protein V2 |
| chr24_+_2495197 | 3.25 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr11_+_12720171 | 3.23 |
ENSDART00000135761
ENSDART00000122812 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr19_+_19489588 | 3.20 |
ENSDART00000166755
|
tril
|
TLR4 interactor with leucine-rich repeats |
| chr13_+_25422009 | 3.16 |
ENSDART00000057686
|
calhm2
|
calcium homeostasis modulator 2 |
| chr4_-_17669881 | 3.08 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr7_+_23577723 | 3.07 |
ENSDART00000049885
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr3_-_39363065 | 3.06 |
ENSDART00000155894
|
arhgap23a
|
Rho GTPase activating protein 23a |
| chr17_+_29345606 | 3.05 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr25_-_31118923 | 2.98 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr2_-_32237916 | 2.95 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr3_-_53486169 | 2.95 |
ENSDART00000115243
|
soul5
|
heme-binding protein soul5 |
| chr14_+_31865099 | 2.79 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr5_-_51998708 | 2.78 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr23_-_21533949 | 2.73 |
ENSDART00000189010
|
rcc2
|
regulator of chromosome condensation 2 |
| chr2_+_36646451 | 2.72 |
ENSDART00000039174
|
klhl6
|
kelch-like family member 6 |
| chr21_+_22878991 | 2.67 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr10_+_7102461 | 2.65 |
ENSDART00000187539
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr6_+_36821621 | 2.64 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr21_-_18086114 | 2.61 |
ENSDART00000143987
|
vav2
|
vav 2 guanine nucleotide exchange factor |
| chr16_-_6849754 | 2.60 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr24_-_26622423 | 2.57 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr16_+_2843428 | 2.52 |
ENSDART00000149485
|
clec3ba
|
C-type lectin domain family 3, member Ba |
| chr19_+_47311020 | 2.51 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr19_+_32166702 | 2.49 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr3_+_4213864 | 2.48 |
ENSDART00000059613
|
si:dkeyp-52c3.5
|
si:dkeyp-52c3.5 |
| chr24_-_25166416 | 2.47 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr6_+_51824596 | 2.46 |
ENSDART00000149003
|
rspo4
|
R-spondin 4 |
| chr6_-_10809546 | 2.42 |
ENSDART00000151661
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr16_-_17300030 | 2.41 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr7_-_2090594 | 2.33 |
ENSDART00000183166
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr10_-_2971407 | 2.31 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr8_-_29706882 | 2.30 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr6_-_1761256 | 2.30 |
ENSDART00000171313
|
orc4
|
origin recognition complex, subunit 4 |
| chr22_-_5933844 | 2.30 |
ENSDART00000163370
ENSDART00000189331 |
si:rp71-36a1.2
|
si:rp71-36a1.2 |
| chr22_-_6884981 | 2.26 |
ENSDART00000124219
|
FO904898.4
|
|
| chr19_+_22216778 | 2.21 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr19_+_5315987 | 2.20 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr5_-_31904562 | 2.19 |
ENSDART00000140640
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr13_+_25421531 | 2.15 |
ENSDART00000158093
|
calhm2
|
calcium homeostasis modulator 2 |
| chr3_-_3731563 | 2.15 |
ENSDART00000180978
|
BX908804.2
|
|
| chr7_-_19332293 | 2.11 |
ENSDART00000169668
ENSDART00000137575 ENSDART00000090406 |
dock11
|
dedicator of cytokinesis 11 |
| chr11_+_34921492 | 2.11 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr7_+_34290051 | 2.10 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr23_+_19977120 | 2.05 |
ENSDART00000089342
|
cfap126
|
cilia and flagella associated protein 126 |
| chr10_-_20537000 | 2.04 |
ENSDART00000064603
ENSDART00000135341 |
plpp5
|
phospholipid phosphatase 5 |
| chr11_+_30647545 | 2.02 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr23_+_26079467 | 2.01 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr5_-_22082918 | 1.98 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr9_-_27410597 | 1.95 |
ENSDART00000135652
ENSDART00000042297 |
kdelc1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr5_+_31480342 | 1.93 |
ENSDART00000098197
|
si:dkey-220k22.1
|
si:dkey-220k22.1 |
| chr1_+_49878000 | 1.91 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr9_-_44983666 | 1.88 |
ENSDART00000149704
|
itgb2
|
integrin, beta 2 |
| chr20_+_20731633 | 1.88 |
ENSDART00000191952
ENSDART00000165224 |
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr16_+_42830152 | 1.87 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr18_+_402048 | 1.86 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr24_-_1303935 | 1.85 |
ENSDART00000159212
ENSDART00000159267 ENSDART00000164904 |
nrp1a
|
neuropilin 1a |
| chr16_+_42829735 | 1.83 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr20_-_40451115 | 1.81 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr6_-_50704689 | 1.80 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr3_+_4047789 | 1.78 |
ENSDART00000148498
|
si:dkeyp-52c3.5
|
si:dkeyp-52c3.5 |
| chr25_-_34632050 | 1.78 |
ENSDART00000170671
|
megf8
|
multiple EGF-like-domains 8 |
| chr18_+_17959681 | 1.76 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr5_-_24124118 | 1.76 |
ENSDART00000051550
|
capga
|
capping protein (actin filament), gelsolin-like a |
| chr12_-_7806007 | 1.75 |
ENSDART00000190359
|
ank3b
|
ankyrin 3b |
| chr25_+_15841670 | 1.73 |
ENSDART00000049992
|
syt9b
|
synaptotagmin IXb |
| chr2_-_42704863 | 1.66 |
ENSDART00000140159
|
myo10
|
myosin X |
| chr17_+_38255105 | 1.65 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr7_-_56606752 | 1.65 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr24_-_37877743 | 1.62 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr16_-_26232411 | 1.60 |
ENSDART00000139355
|
arhgef1b
|
Rho guanine nucleotide exchange factor (GEF) 1b |
| chr20_-_38446891 | 1.57 |
ENSDART00000192013
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr2_+_31671545 | 1.56 |
ENSDART00000145446
|
ackr4a
|
atypical chemokine receptor 4a |
| chr7_-_41403022 | 1.55 |
ENSDART00000174285
|
BX322787.1
|
|
| chr17_-_19344999 | 1.52 |
ENSDART00000138315
|
gsc
|
goosecoid |
| chr24_+_41610068 | 1.48 |
ENSDART00000159507
|
CABZ01044108.1
|
|
| chr14_-_24001825 | 1.42 |
ENSDART00000130704
|
il4
|
interleukin 4 |
| chr2_-_16449504 | 1.42 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr6_+_1787160 | 1.42 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr21_+_42708621 | 1.41 |
ENSDART00000158392
|
si:ch1073-204b8.1
|
si:ch1073-204b8.1 |
| chr4_-_9609634 | 1.37 |
ENSDART00000067188
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr9_-_12424231 | 1.34 |
ENSDART00000188952
|
zgc:162707
|
zgc:162707 |
| chr10_+_40836378 | 1.34 |
ENSDART00000085792
|
trim69
|
tripartite motif containing 69 |
| chr5_-_21888368 | 1.33 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr5_-_39805620 | 1.32 |
ENSDART00000137801
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr2_-_44746723 | 1.29 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium chain family member 3 |
| chr7_-_25895189 | 1.29 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr7_+_5494565 | 1.28 |
ENSDART00000135271
|
MYADM
|
si:dkeyp-67a8.2 |
| chr7_-_52153105 | 1.27 |
ENSDART00000174378
|
CT737190.1
|
|
| chr19_+_7627070 | 1.27 |
ENSDART00000151078
ENSDART00000131324 |
pygo2
|
pygopus homolog 2 (Drosophila) |
| chr10_+_24445698 | 1.26 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr8_+_28358161 | 1.25 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr12_-_34716037 | 1.23 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr7_-_71837213 | 1.23 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr6_-_9707599 | 1.23 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr10_+_28587446 | 1.21 |
ENSDART00000030138
ENSDART00000137508 |
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr15_+_33989181 | 1.20 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr5_-_39805874 | 1.15 |
ENSDART00000176202
ENSDART00000191683 |
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr7_+_72003301 | 1.14 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr7_-_31794476 | 1.13 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr24_-_1303553 | 1.02 |
ENSDART00000190984
|
nrp1a
|
neuropilin 1a |
| chr20_+_405811 | 1.01 |
ENSDART00000149311
|
gpr63
|
G protein-coupled receptor 63 |
| chr19_-_1895832 | 1.01 |
ENSDART00000097530
ENSDART00000192739 ENSDART00000181795 |
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr10_-_24371312 | 0.98 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr5_-_14509137 | 0.97 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr20_-_14875308 | 0.97 |
ENSDART00000141290
|
dnm3a
|
dynamin 3a |
| chr22_+_25590391 | 0.97 |
ENSDART00000178133
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr3_+_13242690 | 0.94 |
ENSDART00000157097
|
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr13_-_33170733 | 0.94 |
ENSDART00000057382
|
fbln5
|
fibulin 5 |
| chr16_-_26437668 | 0.93 |
ENSDART00000142056
|
megf8
|
multiple EGF-like-domains 8 |
| chr24_-_37877978 | 0.92 |
ENSDART00000128574
|
tmem204
|
transmembrane protein 204 |
| chr3_-_34586403 | 0.92 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr2_+_35732652 | 0.92 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr19_+_20178978 | 0.91 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr21_-_20342096 | 0.89 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr12_+_27055900 | 0.87 |
ENSDART00000153198
|
fbrs
|
fibrosin |
| chr8_-_22558773 | 0.84 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr18_-_46369516 | 0.84 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr20_+_23408970 | 0.80 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr23_-_19225709 | 0.75 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr7_-_6754012 | 0.75 |
ENSDART00000113658
|
zgc:55262
|
zgc:55262 |
| chr3_+_32129632 | 0.69 |
ENSDART00000174522
|
zgc:109934
|
zgc:109934 |
| chr16_+_8716800 | 0.68 |
ENSDART00000124693
ENSDART00000181961 |
cabz01093075.1
|
cabz01093075.1 |
| chr19_+_27339848 | 0.64 |
ENSDART00000052355
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr8_+_7033049 | 0.63 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr5_-_19861766 | 0.63 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr10_-_11761927 | 0.61 |
ENSDART00000138748
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr21_-_5879897 | 0.60 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr5_-_24231139 | 0.57 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr20_+_14968031 | 0.55 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
| chr13_-_36663358 | 0.55 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr15_-_16070731 | 0.55 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr19_-_3482138 | 0.55 |
ENSDART00000168139
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr1_-_21163532 | 0.54 |
ENSDART00000087670
ENSDART00000188893 |
si:dkey-253i9.4
|
si:dkey-253i9.4 |
| chr22_+_17606863 | 0.54 |
ENSDART00000035670
|
polr2eb
|
polymerase (RNA) II (DNA directed) polypeptide E, b |
| chr16_+_26601364 | 0.53 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr1_-_9885320 | 0.51 |
ENSDART00000054843
ENSDART00000138630 |
zgc:56409
|
zgc:56409 |
| chr18_+_48953963 | 0.50 |
ENSDART00000158104
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr20_+_6590220 | 0.50 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr17_-_8727699 | 0.49 |
ENSDART00000049236
ENSDART00000149505 ENSDART00000148619 ENSDART00000149668 ENSDART00000148827 |
ctbp2a
|
C-terminal binding protein 2a |
| chr14_+_48964628 | 0.49 |
ENSDART00000105427
|
mif
|
macrophage migration inhibitory factor |
| chr9_-_12424791 | 0.48 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr4_+_77973876 | 0.44 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr2_-_37043540 | 0.44 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb18+zbtb42
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.6 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 1.7 | 5.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) anterior lateral line neuromast hair cell development(GO:0035676) |
| 1.2 | 4.6 | GO:0044003 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.9 | 6.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.8 | 5.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.8 | 3.9 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.7 | 3.7 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.7 | 7.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.7 | 3.4 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.6 | 4.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.6 | 2.9 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.5 | 1.4 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.5 | 3.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.4 | 4.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.4 | 2.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.4 | 1.9 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.3 | 10.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.3 | 1.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.3 | 1.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 0.8 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 1.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 3.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 5.6 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.2 | 2.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 3.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 10.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.2 | 1.8 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.2 | 0.6 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 2.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.8 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 1.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 4.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 3.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.5 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 3.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 10.0 | GO:0060348 | bone development(GO:0060348) |
| 0.1 | 2.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 4.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 4.5 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.4 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 1.6 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 2.9 | GO:1903039 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.9 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 1.0 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 1.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.7 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 2.0 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 5.5 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.1 | 2.5 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 0.5 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 2.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.7 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 2.6 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 1.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 2.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 13.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 1.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 2.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 4.2 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 1.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 6.6 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.9 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 2.7 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 5.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 2.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 2.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 3.1 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 2.0 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 2.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.2 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.8 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 8.2 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 3.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 1.5 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 2.3 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.0 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 4.0 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 1.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 3.2 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.6 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.6 | 2.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 4.0 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 2.0 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.3 | 6.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.3 | 6.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.3 | 4.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 1.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 4.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 2.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 3.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 2.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 25.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 6.1 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 2.7 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.4 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.0 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 65.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.9 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 2.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 6.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 4.4 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.3 | 10.2 | GO:0036122 | BMP binding(GO:0036122) |
| 1.0 | 11.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.8 | 5.6 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.7 | 5.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.6 | 2.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.5 | 4.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.4 | 7.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.4 | 1.2 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.3 | 4.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.3 | 1.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 4.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.3 | 6.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 2.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 4.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 4.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 2.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 4.0 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.2 | 3.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 3.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 0.6 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 1.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 3.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 16.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 3.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 3.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.6 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 11.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.3 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 2.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 7.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 3.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 2.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 11.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 4.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 5.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.8 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.5 | 11.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.5 | 5.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.4 | 3.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 1.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 3.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 2.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 7.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 2.6 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 4.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 12.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.6 | 6.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.5 | 3.0 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.4 | 3.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 6.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.3 | 4.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 7.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 11.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 4.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 4.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 3.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |