Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
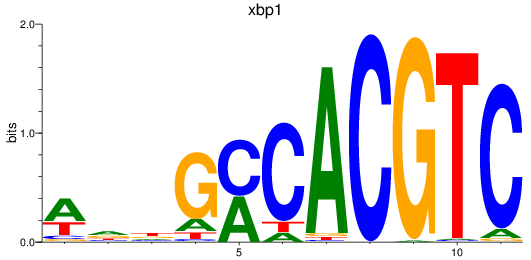
Results for xbp1
Z-value: 1.52
Transcription factors associated with xbp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
xbp1
|
ENSDARG00000035622 | X-box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| xbp1 | dr11_v1_chr5_-_15948833_15948920 | 0.77 | 2.1e-19 | Click! |
Activity profile of xbp1 motif
Sorted Z-values of xbp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_23984755 | 28.56 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr7_+_10610791 | 18.71 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr16_+_27224369 | 15.99 |
ENSDART00000136980
|
sec61b
|
Sec61 translocon beta subunit |
| chr21_+_10702031 | 15.67 |
ENSDART00000102304
|
lman1
|
lectin, mannose-binding, 1 |
| chr21_+_10701834 | 15.15 |
ENSDART00000192473
|
lman1
|
lectin, mannose-binding, 1 |
| chr8_-_14554785 | 14.50 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr24_+_14214831 | 14.26 |
ENSDART00000004664
|
tram1
|
translocation associated membrane protein 1 |
| chr16_+_23984179 | 13.32 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr24_-_42090635 | 13.02 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr12_-_990149 | 12.35 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr22_+_38194151 | 10.69 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr1_-_23308225 | 10.64 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr16_-_46660680 | 10.62 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr6_+_6779114 | 10.62 |
ENSDART00000163493
ENSDART00000143359 |
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr10_-_22150419 | 10.43 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr11_+_44503774 | 10.10 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr19_-_11238620 | 9.20 |
ENSDART00000148697
ENSDART00000149169 |
ssr2
|
signal sequence receptor, beta |
| chr6_+_13506841 | 8.64 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr8_+_5024468 | 8.61 |
ENSDART00000030938
|
adra1aa
|
adrenoceptor alpha 1Aa |
| chr15_-_32383340 | 8.36 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr4_+_25651720 | 8.09 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr1_-_45049603 | 7.78 |
ENSDART00000023336
|
rps6
|
ribosomal protein S6 |
| chr23_-_18572685 | 7.76 |
ENSDART00000047175
|
ssr4
|
signal sequence receptor, delta |
| chr23_-_18572521 | 7.49 |
ENSDART00000138599
|
ssr4
|
signal sequence receptor, delta |
| chr6_-_40071899 | 7.40 |
ENSDART00000034730
|
sec61a1
|
Sec61 translocon alpha 1 subunit |
| chr2_-_42492201 | 7.32 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr7_+_44715224 | 7.25 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr8_+_11471350 | 7.09 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr4_-_13613148 | 7.02 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr6_-_23931442 | 7.00 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr25_+_5983430 | 6.87 |
ENSDART00000074814
|
ppib
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr4_-_17725008 | 6.81 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr8_+_47219107 | 6.73 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr6_-_42388608 | 6.72 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr19_+_18903533 | 6.66 |
ENSDART00000157523
ENSDART00000166562 |
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr10_+_39212898 | 6.39 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr12_+_31783066 | 6.33 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr4_-_19693978 | 6.28 |
ENSDART00000100974
ENSDART00000040405 |
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr3_-_25377163 | 6.15 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr4_+_68562464 | 6.07 |
ENSDART00000192954
|
BX548011.4
|
|
| chr20_-_438646 | 6.06 |
ENSDART00000009196
|
ufl1
|
UFM1-specific ligase 1 |
| chr5_+_24543862 | 6.06 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase H+ transporting V0 subunit a2b |
| chr19_+_43119698 | 5.84 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr5_+_2002804 | 5.83 |
ENSDART00000064088
|
vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr20_-_30377221 | 5.73 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr19_-_30404096 | 5.69 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr17_+_23311377 | 5.53 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr2_-_55298075 | 5.48 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr1_-_53277187 | 5.34 |
ENSDART00000131520
|
znf330
|
zinc finger protein 330 |
| chr24_+_42074143 | 5.32 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr17_+_15213496 | 5.12 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr15_-_32383529 | 5.02 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr19_+_14113886 | 4.97 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr1_-_53277413 | 4.91 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr2_-_7757273 | 4.90 |
ENSDART00000136074
|
b3gnt5b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5b |
| chr1_-_44484 | 4.89 |
ENSDART00000171547
ENSDART00000164075 ENSDART00000168091 |
tmem39a
|
transmembrane protein 39A |
| chr22_-_817479 | 4.81 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr10_+_39212601 | 4.81 |
ENSDART00000168778
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr17_+_53156530 | 4.73 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr14_-_45464112 | 4.69 |
ENSDART00000173209
|
ehd1a
|
EH-domain containing 1a |
| chr18_-_34170918 | 4.63 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr4_+_13568469 | 4.55 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr22_-_17781213 | 4.32 |
ENSDART00000137984
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr10_-_3427589 | 4.32 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr22_-_10152629 | 4.31 |
ENSDART00000144209
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr18_-_34171280 | 4.31 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr5_+_65086856 | 4.11 |
ENSDART00000169209
ENSDART00000162409 |
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr22_+_31059919 | 4.11 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr16_+_53603945 | 3.90 |
ENSDART00000083513
|
sacm1lb
|
SAC1 like phosphatidylinositide phosphatase b |
| chr4_-_779796 | 3.82 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr5_+_66220601 | 3.70 |
ENSDART00000129903
|
mfsd10
|
major facilitator superfamily domain containing 10 |
| chr12_+_1592146 | 3.69 |
ENSDART00000184575
ENSDART00000192902 |
SLC39A11
|
solute carrier family 39 member 11 |
| chr5_-_63644938 | 3.69 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr9_+_21165017 | 3.57 |
ENSDART00000145933
ENSDART00000142985 |
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr5_+_65087226 | 3.56 |
ENSDART00000183187
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr25_+_3759553 | 3.54 |
ENSDART00000180601
ENSDART00000055845 ENSDART00000157050 ENSDART00000153905 |
thoc5
|
THO complex 5 |
| chr6_-_32087108 | 3.43 |
ENSDART00000150934
ENSDART00000130627 |
alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr20_-_40758410 | 3.37 |
ENSDART00000183031
|
cx34.5
|
connexin 34.5 |
| chr18_-_16924221 | 3.34 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr5_+_69868911 | 3.33 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr19_-_29832876 | 3.25 |
ENSDART00000005119
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr9_-_27748868 | 3.22 |
ENSDART00000190306
|
tbccd1
|
TBCC domain containing 1 |
| chr25_-_3759322 | 3.22 |
ENSDART00000088077
|
zgc:158398
|
zgc:158398 |
| chr2_-_22363460 | 3.20 |
ENSDART00000158486
|
selenof
|
selenoprotein F |
| chr20_+_22799857 | 3.20 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr6_+_47843760 | 3.19 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr24_-_26485098 | 3.16 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr25_-_774350 | 3.14 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr3_-_18384501 | 3.10 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr25_-_3034668 | 3.08 |
ENSDART00000053405
|
scamp2
|
secretory carrier membrane protein 2 |
| chr2_+_36608387 | 3.07 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr2_+_37176814 | 3.03 |
ENSDART00000048277
ENSDART00000176827 |
copa
|
coatomer protein complex, subunit alpha |
| chr15_-_29326254 | 3.02 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr14_+_20351 | 2.98 |
ENSDART00000051893
|
stx18
|
syntaxin 18 |
| chr23_+_43638982 | 2.97 |
ENSDART00000168646
|
slc10a7
|
solute carrier family 10, member 7 |
| chr1_-_9486214 | 2.92 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr25_-_37262220 | 2.91 |
ENSDART00000153789
ENSDART00000155182 |
rfwd3
|
ring finger and WD repeat domain 3 |
| chr16_-_26676685 | 2.90 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr12_-_35582683 | 2.85 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr6_-_35052145 | 2.84 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr6_+_3864040 | 2.82 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr3_-_34586403 | 2.81 |
ENSDART00000151515
|
sept9a
|
septin 9a |
| chr8_-_31701157 | 2.81 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr5_+_65086668 | 2.75 |
ENSDART00000183746
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr24_-_11069237 | 2.74 |
ENSDART00000127398
|
CR753886.1
|
|
| chr18_-_21746421 | 2.69 |
ENSDART00000188809
|
pskh1
|
protein serine kinase H1 |
| chr9_+_54695981 | 2.68 |
ENSDART00000183605
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr23_+_42819221 | 2.67 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr11_-_270210 | 2.66 |
ENSDART00000005217
ENSDART00000172779 |
alas1
|
aminolevulinate, delta-, synthase 1 |
| chr3_+_29510818 | 2.65 |
ENSDART00000055407
ENSDART00000193743 ENSDART00000123619 |
rac2
|
Rac family small GTPase 2 |
| chr12_+_30788912 | 2.64 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr5_+_22791686 | 2.63 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr1_-_53277660 | 2.61 |
ENSDART00000014232
|
znf330
|
zinc finger protein 330 |
| chr7_-_69352424 | 2.59 |
ENSDART00000170714
|
ap1g1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr16_+_35918463 | 2.59 |
ENSDART00000160608
|
sh3d21
|
SH3 domain containing 21 |
| chr6_+_58289335 | 2.56 |
ENSDART00000177399
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr6_+_23026714 | 2.53 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr3_+_28860283 | 2.51 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr5_+_58550291 | 2.50 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr5_-_28968964 | 2.46 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr5_+_21211135 | 2.46 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr16_-_42750295 | 2.42 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr17_+_23730089 | 2.42 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr1_-_9485939 | 2.39 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr10_+_16165533 | 2.32 |
ENSDART00000065045
|
prrc1
|
proline-rich coiled-coil 1 |
| chr2_-_9527129 | 2.30 |
ENSDART00000157422
ENSDART00000004398 |
cope
|
coatomer protein complex, subunit epsilon |
| chr9_+_25840720 | 2.30 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr10_-_44305180 | 2.28 |
ENSDART00000187552
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr10_-_34915886 | 2.26 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr15_+_35933094 | 2.21 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr2_-_1227221 | 2.20 |
ENSDART00000130897
|
ABCF3
|
ATP binding cassette subfamily F member 3 |
| chr5_+_6672870 | 2.17 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr15_-_18115540 | 2.14 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr19_+_1414604 | 2.13 |
ENSDART00000159024
|
btr29
|
bloodthirsty-related gene family, member 29 |
| chr6_-_52348562 | 2.13 |
ENSDART00000142565
ENSDART00000145369 ENSDART00000016890 |
eif6
|
eukaryotic translation initiation factor 6 |
| chr14_+_9581896 | 2.12 |
ENSDART00000114563
|
tmem129
|
transmembrane protein 129, E3 ubiquitin protein ligase |
| chr18_+_6706140 | 2.10 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr20_+_22799641 | 2.09 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr11_-_6420917 | 2.03 |
ENSDART00000193717
|
FO681393.2
|
|
| chr22_-_11078988 | 2.00 |
ENSDART00000126664
ENSDART00000006927 |
use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr25_+_34862225 | 2.00 |
ENSDART00000149782
|
CHST6
|
zgc:194879 |
| chr9_-_56399699 | 1.98 |
ENSDART00000170281
|
rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr3_+_53773256 | 1.93 |
ENSDART00000170461
|
col5a3a
|
collagen, type V, alpha 3a |
| chr19_+_26640096 | 1.87 |
ENSDART00000067793
|
ints3
|
integrator complex subunit 3 |
| chr4_-_5912951 | 1.87 |
ENSDART00000169439
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr11_+_6431133 | 1.87 |
ENSDART00000190742
|
FO904935.1
|
|
| chr11_+_30647545 | 1.86 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr13_-_9875538 | 1.84 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr12_-_3773869 | 1.83 |
ENSDART00000092983
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr5_-_30516646 | 1.83 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr16_-_54978981 | 1.83 |
ENSDART00000154023
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr7_-_65114067 | 1.82 |
ENSDART00000110614
ENSDART00000098277 |
tmem41b
|
transmembrane protein 41B |
| chr3_-_36209936 | 1.80 |
ENSDART00000175208
|
csnk1da
|
casein kinase 1, delta a |
| chr6_-_35052388 | 1.78 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr15_-_14193659 | 1.78 |
ENSDART00000171197
|
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr3_-_180860 | 1.77 |
ENSDART00000059956
ENSDART00000192506 |
kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr12_-_16452200 | 1.77 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr18_-_7481036 | 1.71 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr20_+_42537768 | 1.66 |
ENSDART00000134066
ENSDART00000153434 |
si:dkeyp-93d12.1
|
si:dkeyp-93d12.1 |
| chr13_-_47403154 | 1.65 |
ENSDART00000114318
|
bcl2l11
|
BCL2 like 11 |
| chr22_-_20386982 | 1.64 |
ENSDART00000089015
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr12_+_30789611 | 1.63 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr1_-_156375 | 1.62 |
ENSDART00000160221
|
pcid2
|
PCI domain containing 2 |
| chr2_+_43920461 | 1.61 |
ENSDART00000123673
|
si:ch211-195h23.3
|
si:ch211-195h23.3 |
| chr5_-_56964547 | 1.60 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr5_+_20453874 | 1.58 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr1_+_29798140 | 1.58 |
ENSDART00000019743
|
tm9sf2
|
transmembrane 9 superfamily member 2 |
| chr3_-_34180364 | 1.56 |
ENSDART00000151819
ENSDART00000003133 |
yipf2
|
Yip1 domain family, member 2 |
| chr10_+_37173029 | 1.52 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr9_+_23825440 | 1.50 |
ENSDART00000138470
|
ints6
|
integrator complex subunit 6 |
| chr18_-_18587745 | 1.50 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr6_+_49722970 | 1.45 |
ENSDART00000155934
ENSDART00000154738 |
stx16
|
syntaxin 16 |
| chr8_-_44463985 | 1.44 |
ENSDART00000016845
|
mhc1lba
|
major histocompatibility complex class I LBA |
| chr10_-_26225548 | 1.44 |
ENSDART00000132019
ENSDART00000079194 |
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr9_+_54237100 | 1.44 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr10_-_2682198 | 1.42 |
ENSDART00000183727
|
pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr15_+_41815703 | 1.39 |
ENSDART00000059508
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr11_+_7276824 | 1.37 |
ENSDART00000185992
|
si:ch73-238c9.1
|
si:ch73-238c9.1 |
| chr25_-_34512102 | 1.37 |
ENSDART00000087450
|
klf13
|
Kruppel-like factor 13 |
| chr2_+_20926202 | 1.36 |
ENSDART00000178880
|
pla2g4aa
|
phospholipase A2, group IVAa (cytosolic, calcium-dependent) |
| chr12_-_4206869 | 1.35 |
ENSDART00000106572
|
si:dkey-32n7.9
|
si:dkey-32n7.9 |
| chr20_-_26929685 | 1.29 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr6_-_40885496 | 1.27 |
ENSDART00000189857
|
sirt4
|
sirtuin 4 |
| chr14_+_1014109 | 1.24 |
ENSDART00000157945
|
f8
|
coagulation factor VIII, procoagulant component |
| chr12_-_35582521 | 1.20 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr16_+_38159758 | 1.14 |
ENSDART00000058666
ENSDART00000112165 |
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr24_-_33291784 | 1.13 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr21_-_22693730 | 1.08 |
ENSDART00000158342
|
gig2d
|
grass carp reovirus (GCRV)-induced gene 2d |
| chr25_-_1687481 | 1.06 |
ENSDART00000163553
ENSDART00000179460 |
mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr7_+_18100996 | 1.06 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr15_-_29387446 | 1.01 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr5_-_41841675 | 1.00 |
ENSDART00000141683
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr17_-_24866727 | 0.97 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr16_+_6727195 | 0.95 |
ENSDART00000148625
|
znf407
|
zinc finger protein 407 |
| chr4_-_77979432 | 0.93 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr13_-_38088627 | 0.93 |
ENSDART00000175268
|
CT027676.1
|
|
| chr4_+_77957611 | 0.92 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr19_-_30403922 | 0.92 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr9_+_20526472 | 0.92 |
ENSDART00000166457
ENSDART00000141775 |
trim45
|
tripartite motif containing 45 |
| chr17_-_24866964 | 0.91 |
ENSDART00000190601
ENSDART00000192547 |
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
Network of associatons between targets according to the STRING database.
First level regulatory network of xbp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 44.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 3.1 | 18.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 2.5 | 17.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 2.0 | 38.0 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 1.9 | 5.8 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 1.8 | 7.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 1.7 | 10.4 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 1.7 | 9.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.4 | 8.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 1.4 | 7.0 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 1.4 | 5.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 1.4 | 4.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.2 | 8.6 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 1.2 | 6.1 | GO:1903573 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 1.2 | 59.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 1.0 | 14.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 1.0 | 5.0 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.9 | 4.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.8 | 4.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.7 | 9.7 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.7 | 2.1 | GO:1902626 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.7 | 6.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.6 | 2.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.6 | 3.2 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.6 | 11.2 | GO:0043687 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.6 | 10.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.5 | 1.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.5 | 2.7 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.5 | 1.6 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.5 | 6.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.5 | 1.9 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 1.8 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.5 | 3.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.4 | 6.7 | GO:0035999 | methionine biosynthetic process(GO:0009086) tetrahydrofolate interconversion(GO:0035999) |
| 0.4 | 3.1 | GO:1902902 | negative regulation of autophagosome maturation(GO:1901097) negative regulation of autophagosome assembly(GO:1902902) |
| 0.4 | 3.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.4 | 6.1 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.4 | 2.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.4 | 1.4 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.3 | 3.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 5.3 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.3 | 2.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 8.9 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.3 | 1.4 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.3 | 0.8 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 5.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 10.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.3 | 5.5 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.2 | 0.7 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 0.5 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.2 | 6.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 2.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.2 | 0.8 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 0.9 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 1.4 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.2 | 2.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 3.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 2.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 13.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 2.6 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 5.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 3.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 14.5 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 3.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 4.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 5.5 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 8.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 2.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 5.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.4 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 5.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 2.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 7.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 1.9 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 4.3 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.1 | 2.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 2.1 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 3.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 5.7 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.1 | 0.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 7.8 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 1.1 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 6.3 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 3.3 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 1.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 2.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 2.1 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 6.8 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.1 | 0.7 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.4 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 2.5 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 2.3 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 1.1 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 1.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 7.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 2.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.9 | GO:0043491 | protein kinase B signaling(GO:0043491) |
| 0.0 | 0.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 3.1 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.5 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.4 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 1.9 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 1.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 1.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.5 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 1.0 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 2.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 30.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 4.2 | 41.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 1.8 | 26.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 1.0 | 26.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.8 | 3.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.8 | 6.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.8 | 2.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.7 | 35.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.6 | 4.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 6.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.5 | 2.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.5 | 3.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.5 | 4.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 1.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.4 | 1.8 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.4 | 3.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.4 | 3.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.4 | 3.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 2.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 2.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.3 | 5.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 1.6 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.3 | 1.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 5.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.3 | 9.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.3 | 5.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 5.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.2 | 14.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.2 | 4.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 1.5 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.2 | 113.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.2 | 5.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 14.2 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 33.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.1 | 2.8 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 5.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 11.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 12.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 4.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 2.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 3.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 4.8 | 14.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 3.7 | 18.7 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 2.8 | 11.2 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 2.4 | 30.8 | GO:0005537 | mannose binding(GO:0005537) |
| 2.2 | 8.6 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 2.0 | 6.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 1.6 | 4.9 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.5 | 8.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 1.5 | 10.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.4 | 6.8 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 1.3 | 5.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 1.3 | 3.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 1.1 | 8.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.9 | 14.1 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.9 | 4.6 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.9 | 9.9 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.9 | 2.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.9 | 4.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.7 | 10.7 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.7 | 7.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.6 | 3.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.6 | 1.8 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.5 | 2.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.5 | 1.9 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.4 | 6.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 2.4 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 6.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.4 | 5.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 6.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.4 | 3.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 2.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.3 | 8.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.3 | 1.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.3 | 5.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 6.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.2 | 1.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 5.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 9.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 3.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.2 | 2.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 4.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 1.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 6.3 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 1.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 2.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 5.1 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.1 | 6.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 49.3 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.1 | 2.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 10.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 1.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 8.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 4.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 13.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 3.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 2.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 13.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 7.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 2.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.7 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 3.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.4 | GO:0019904 | protein domain specific binding(GO:0019904) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 37.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.4 | 8.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 3.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 1.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 10.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 2.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 3.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 3.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 2.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 5.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 2.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 5.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 7.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 2.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 41.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.8 | 39.0 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.8 | 17.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.8 | 7.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.6 | 83.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.5 | 5.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.4 | 4.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.4 | 6.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.4 | 22.3 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.3 | 5.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 8.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 2.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 5.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 3.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 3.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.3 | 2.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 7.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 2.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.0 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 6.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 18.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 5.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.1 | REACTOME INTERFERON SIGNALING | Genes involved in Interferon Signaling |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |