Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
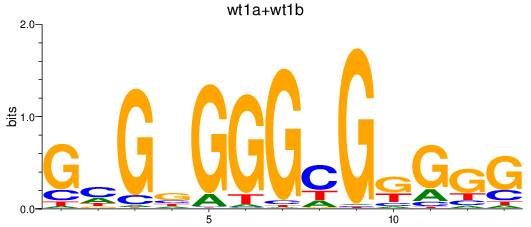
Results for wt1a+wt1b
Z-value: 1.44
Transcription factors associated with wt1a+wt1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
wt1b
|
ENSDARG00000007990 | WT1 transcription factor b |
|
wt1a
|
ENSDARG00000031420 | WT1 transcription factor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| wt1b | dr11_v1_chr18_-_45617146_45617146 | -0.25 | 1.5e-02 | Click! |
| wt1a | dr11_v1_chr25_-_15214161_15214161 | -0.15 | 1.6e-01 | Click! |
Activity profile of wt1a+wt1b motif
Sorted Z-values of wt1a+wt1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_48111285 | 12.67 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr7_-_10606 | 11.97 |
ENSDART00000192650
ENSDART00000186761 |
FO704772.2
|
|
| chr2_+_59015878 | 11.81 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr13_+_22480857 | 10.75 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr3_+_32526799 | 10.71 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr2_-_22535 | 10.60 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr3_-_1190132 | 10.46 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr2_-_44282796 | 9.94 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr3_+_32526263 | 9.94 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr1_-_59232267 | 9.22 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr18_+_5549672 | 8.26 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr25_+_150570 | 7.99 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr22_+_396840 | 7.76 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr21_-_5056812 | 7.63 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr17_-_125091 | 7.63 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr24_+_42131564 | 7.61 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr2_+_58221163 | 7.57 |
ENSDART00000157939
|
FO704813.1
|
|
| chr19_+_33701558 | 7.47 |
ENSDART00000147226
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr13_-_31435137 | 7.46 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr9_-_39005317 | 7.29 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr25_-_37501371 | 7.24 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr10_+_32561317 | 6.97 |
ENSDART00000109029
|
map6a
|
microtubule-associated protein 6a |
| chr14_+_2243 | 6.92 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr25_+_34984333 | 6.85 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr23_+_28731379 | 6.68 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr2_-_48753873 | 6.62 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr20_-_14665002 | 6.56 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr4_+_41602 | 6.19 |
ENSDART00000159640
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr11_-_97817 | 6.18 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr19_+_31771270 | 6.04 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr7_-_18168493 | 5.98 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr22_-_193234 | 5.88 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr3_-_62380146 | 5.85 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr2_+_24304854 | 5.68 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr19_+_33701366 | 5.57 |
ENSDART00000162517
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr9_-_54840124 | 5.53 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr1_+_57371447 | 5.51 |
ENSDART00000152229
ENSDART00000181077 |
si:dkey-27j5.3
|
si:dkey-27j5.3 |
| chr7_-_57949117 | 5.47 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr3_+_5575313 | 5.43 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr1_-_14234076 | 5.39 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr16_-_6821927 | 5.37 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr1_-_14233815 | 5.34 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr20_-_54381034 | 5.34 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr3_+_50312422 | 5.23 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr24_+_24831727 | 5.20 |
ENSDART00000080969
|
trim55b
|
tripartite motif containing 55b |
| chr16_-_12953739 | 5.16 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr2_-_9646857 | 5.16 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr6_-_13408680 | 5.13 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr21_-_22114625 | 5.10 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr24_+_42127983 | 5.05 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr19_+_33701734 | 5.01 |
ENSDART00000123270
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr11_-_103136 | 4.99 |
ENSDART00000173308
ENSDART00000162982 |
elmo2
|
engulfment and cell motility 2 |
| chr8_+_8298439 | 4.95 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr2_+_38924975 | 4.90 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr21_-_308852 | 4.88 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr7_-_6592142 | 4.78 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr16_-_44399335 | 4.77 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr11_+_41560792 | 4.71 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr16_-_44349845 | 4.67 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr15_-_33925851 | 4.66 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr14_+_22172047 | 4.64 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr8_+_54284961 | 4.64 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr20_+_52554352 | 4.60 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr17_+_53311618 | 4.59 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr9_-_54001502 | 4.52 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr11_+_40649412 | 4.50 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr14_-_12837052 | 4.46 |
ENSDART00000165004
ENSDART00000043180 |
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr11_+_45461853 | 4.45 |
ENSDART00000173409
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr1_+_54677173 | 4.43 |
ENSDART00000114705
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr23_-_637347 | 4.41 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr18_+_54354 | 4.34 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr8_+_23165749 | 4.34 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr5_+_36614196 | 4.33 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr3_+_1211242 | 4.32 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr25_+_35706493 | 4.30 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr20_+_54356272 | 4.25 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr17_-_20711735 | 4.24 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr14_-_237130 | 4.17 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr23_-_3703569 | 4.10 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr18_-_50845804 | 4.09 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr18_-_15373620 | 4.08 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr14_-_12837432 | 4.07 |
ENSDART00000178444
|
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr1_+_54908895 | 4.05 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr13_+_12739283 | 4.03 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr15_+_1397811 | 4.02 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr7_+_48460239 | 3.97 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr8_+_8166285 | 3.94 |
ENSDART00000147940
|
plxnb3
|
plexin B3 |
| chr6_+_28203 | 3.93 |
ENSDART00000191561
|
CZQB01141835.1
|
|
| chr17_+_53311243 | 3.91 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr13_-_51922290 | 3.90 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr21_+_13861589 | 3.89 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr21_+_11468934 | 3.87 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr6_+_59808677 | 3.86 |
ENSDART00000164469
|
caskb
|
calcium/calmodulin-dependent serine protein kinase b |
| chr5_-_6433577 | 3.86 |
ENSDART00000160439
|
CABZ01037298.1
|
|
| chr20_-_52928541 | 3.82 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr1_+_29858032 | 3.77 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr1_-_25936677 | 3.76 |
ENSDART00000146488
ENSDART00000136321 |
myoz2b
|
myozenin 2b |
| chr5_+_64739762 | 3.73 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr16_-_563235 | 3.72 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr17_-_43466317 | 3.67 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr7_+_74150839 | 3.62 |
ENSDART00000160195
|
ppp1cbl
|
protein phosphatase 1, catalytic subunit, beta isoform, like |
| chr16_+_12240605 | 3.56 |
ENSDART00000060056
|
tpi1b
|
triosephosphate isomerase 1b |
| chr8_-_54223316 | 3.53 |
ENSDART00000018054
|
trh
|
thyrotropin-releasing hormone |
| chr11_+_6819050 | 3.52 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr13_+_25428677 | 3.50 |
ENSDART00000186284
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr16_+_1803462 | 3.48 |
ENSDART00000183974
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr24_+_42132962 | 3.46 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr13_+_421231 | 3.43 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr19_+_63567 | 3.43 |
ENSDART00000165657
ENSDART00000165183 |
zhx2b
|
zinc fingers and homeoboxes 2b |
| chr8_-_9118958 | 3.36 |
ENSDART00000037922
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr18_-_10298162 | 3.35 |
ENSDART00000007520
|
lrrc4.2
|
leucine rich repeat containing 4.2 |
| chr23_+_44732863 | 3.33 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr5_-_72289648 | 3.32 |
ENSDART00000037691
|
tbx5a
|
T-box 5a |
| chr21_+_11468642 | 3.27 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr10_+_44940693 | 3.22 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr12_+_5189776 | 3.21 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr10_-_24343507 | 3.21 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr8_-_22739757 | 3.14 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr4_+_77948970 | 3.11 |
ENSDART00000149636
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr20_-_54462551 | 3.11 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr5_-_25066780 | 3.10 |
ENSDART00000002118
ENSDART00000182575 |
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr12_-_49165751 | 3.09 |
ENSDART00000148491
ENSDART00000112479 |
acadsb
|
acyl-CoA dehydrogenase short/branched chain |
| chr25_-_19395476 | 3.08 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr9_-_2572790 | 2.99 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr5_+_51443009 | 2.99 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr21_-_45878872 | 2.98 |
ENSDART00000029763
|
sap30l
|
sap30-like |
| chr11_+_2198831 | 2.97 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr9_-_2573121 | 2.96 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr15_-_14486534 | 2.94 |
ENSDART00000179368
|
numbl
|
numb homolog (Drosophila)-like |
| chr23_-_19953089 | 2.94 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr6_-_52428826 | 2.93 |
ENSDART00000047399
|
mmp24
|
matrix metallopeptidase 24 |
| chr10_+_37500234 | 2.91 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr12_+_9880493 | 2.88 |
ENSDART00000055019
|
ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr16_-_42152145 | 2.87 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr21_+_103194 | 2.86 |
ENSDART00000162755
|
zer1
|
zyg-11 related, cell cycle regulator |
| chr7_-_73702063 | 2.80 |
ENSDART00000130539
|
BX664721.1
|
|
| chr22_-_600016 | 2.78 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr24_-_25364775 | 2.78 |
ENSDART00000151915
|
ptchd1
|
patched domain containing 1 |
| chr20_-_39219537 | 2.78 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr11_-_44030962 | 2.76 |
ENSDART00000171910
|
FP016005.1
|
|
| chr19_+_233143 | 2.75 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr6_+_58492201 | 2.75 |
ENSDART00000156375
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr11_+_1796426 | 2.68 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr25_-_19395156 | 2.66 |
ENSDART00000155335
|
map1ab
|
microtubule-associated protein 1Ab |
| chr15_-_8517555 | 2.60 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr25_+_35774544 | 2.60 |
ENSDART00000034737
ENSDART00000188162 |
cpne8
|
copine VIII |
| chr16_-_12914288 | 2.59 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr12_+_18606140 | 2.58 |
ENSDART00000161128
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr19_-_19442983 | 2.58 |
ENSDART00000052649
|
sb:cb649
|
sb:cb649 |
| chr23_+_44644911 | 2.55 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr9_-_1434484 | 2.55 |
ENSDART00000093412
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr21_+_38312549 | 2.53 |
ENSDART00000065159
|
zgc:158291
|
zgc:158291 |
| chr20_+_39250673 | 2.50 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr23_+_19564392 | 2.50 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr6_+_31684 | 2.50 |
ENSDART00000188853
ENSDART00000184553 |
CZQB01141835.2
|
|
| chr7_+_10701938 | 2.48 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr6_+_2271559 | 2.47 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr14_+_11103718 | 2.47 |
ENSDART00000161311
|
nexmifb
|
neurite extension and migration factor b |
| chr22_+_12366516 | 2.46 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr9_-_30363770 | 2.46 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr12_-_26430507 | 2.45 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr10_-_35542071 | 2.45 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr16_-_563732 | 2.42 |
ENSDART00000183394
|
irx2a
|
iroquois homeobox 2a |
| chr2_+_3595333 | 2.42 |
ENSDART00000041052
|
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr9_+_51891 | 2.41 |
ENSDART00000163529
|
zgc:158316
|
zgc:158316 |
| chr6_-_58828113 | 2.41 |
ENSDART00000180934
|
kif5ab
|
kinesin family member 5A, b |
| chr19_+_41701660 | 2.39 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr20_-_1268863 | 2.37 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr23_+_9560797 | 2.37 |
ENSDART00000180014
|
adrm1
|
adhesion regulating molecule 1 |
| chr2_-_11912347 | 2.36 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr7_-_51627019 | 2.36 |
ENSDART00000174111
|
nhsl2
|
NHS-like 2 |
| chr2_-_7246338 | 2.34 |
ENSDART00000186735
|
zgc:153115
|
zgc:153115 |
| chr25_-_207214 | 2.33 |
ENSDART00000193448
|
FP236318.3
|
|
| chr5_-_52277643 | 2.32 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr3_-_6767440 | 2.32 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr16_+_10963602 | 2.31 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr15_-_14552101 | 2.30 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr8_+_2656231 | 2.28 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr22_+_465269 | 2.26 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr19_+_30633453 | 2.26 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr10_+_45128375 | 2.26 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr16_+_23600041 | 2.24 |
ENSDART00000186918
|
kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr3_+_7617353 | 2.23 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr2_-_16216568 | 2.21 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_2369849 | 2.20 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr4_-_837768 | 2.19 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr2_+_26303627 | 2.19 |
ENSDART00000040278
|
efna2a
|
ephrin-A2a |
| chr21_+_233271 | 2.12 |
ENSDART00000171440
|
dtwd2
|
DTW domain containing 2 |
| chr24_+_33622769 | 2.11 |
ENSDART00000079342
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr10_-_40939303 | 2.10 |
ENSDART00000134295
|
bmp1b
|
bone morphogenetic protein 1b |
| chr2_+_49232559 | 2.09 |
ENSDART00000154285
ENSDART00000175147 |
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr20_+_23501535 | 2.08 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr5_+_9348284 | 2.08 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr6_-_16406210 | 2.07 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr21_+_37445871 | 2.07 |
ENSDART00000076333
|
pgk1
|
phosphoglycerate kinase 1 |
| chr9_+_19489304 | 2.07 |
ENSDART00000151920
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr23_+_6752828 | 2.06 |
ENSDART00000105179
|
zgc:158254
|
zgc:158254 |
| chr22_-_18112374 | 2.04 |
ENSDART00000191154
|
ncanb
|
neurocan b |
| chr10_-_1718395 | 2.03 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr17_-_22048233 | 2.02 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of wt1a+wt1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.1 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 2.1 | 8.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.6 | 7.8 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 1.5 | 9.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 1.2 | 6.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 1.2 | 3.5 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.1 | 3.3 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 1.1 | 5.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.9 | 3.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.8 | 12.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.7 | 4.8 | GO:0036268 | swimming(GO:0036268) |
| 0.6 | 2.5 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.6 | 10.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.5 | 4.9 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.5 | 3.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.5 | 6.8 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.5 | 2.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.5 | 4.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.5 | 11.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 4.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.5 | 3.8 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.5 | 2.4 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.5 | 2.8 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.5 | 7.7 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.5 | 1.8 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.5 | 4.5 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.5 | 7.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 1.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.4 | 4.7 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.4 | 3.7 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.4 | 8.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 1.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 2.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 1.5 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.3 | 2.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.3 | 8.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.3 | 5.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.3 | 1.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.3 | 1.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.3 | 5.4 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.3 | 1.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.3 | 2.4 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.3 | 8.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 8.0 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.3 | 1.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.3 | 3.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 1.3 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 11.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 2.8 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.2 | 1.4 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.2 | 2.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 | 4.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 2.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 1.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 1.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 2.0 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 1.5 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 1.7 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 4.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 1.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 1.9 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.2 | 2.4 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.2 | 2.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 3.7 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.2 | 1.3 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.2 | 3.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 2.1 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.2 | 3.3 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.8 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.2 | 0.7 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 1.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 3.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.2 | 1.8 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 1.0 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 13.8 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.2 | 0.5 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 2.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 11.5 | GO:0042552 | myelination(GO:0042552) |
| 0.1 | 3.5 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.8 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 2.8 | GO:0050890 | cognition(GO:0050890) |
| 0.1 | 2.5 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 5.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.7 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.4 | GO:0051958 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 4.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 1.9 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.1 | 3.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.7 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 2.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 2.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 3.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 1.0 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.9 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 13.3 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 0.7 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 4.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 4.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.6 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 1.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 2.9 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 5.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 10.7 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.1 | 0.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.2 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 4.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 2.1 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 2.5 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 24.4 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 0.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 4.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.7 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.9 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.9 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 2.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 3.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.8 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 6.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.7 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.9 | GO:0031577 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 2.3 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 3.0 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 5.3 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 1.0 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 4.3 | GO:0060548 | negative regulation of cell death(GO:0060548) |
| 0.0 | 2.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 8.4 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 7.3 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 1.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.1 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 5.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 2.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.7 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 1.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 4.5 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.2 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.9 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 2.5 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.5 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.7 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 1.7 | 10.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.9 | 15.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.8 | 10.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.7 | 4.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 2.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.5 | 7.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.5 | 7.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 2.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.4 | 8.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.4 | 11.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.4 | 2.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 4.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.4 | 16.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.4 | 1.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.4 | 4.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 3.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 5.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 3.3 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 4.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 3.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 10.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 3.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 2.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 3.8 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 4.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 5.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 3.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 8.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 7.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 6.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.1 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 2.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 6.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 4.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 5.5 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 12.3 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 8.1 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 6.7 | GO:0099503 | exocytic vesicle(GO:0070382) secretory vesicle(GO:0099503) |
| 0.0 | 0.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 2.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 2.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 6.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 13.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 6.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.4 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 2.5 | 12.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 2.1 | 8.3 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 1.3 | 3.8 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.9 | 3.6 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.8 | 2.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.6 | 3.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.6 | 8.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.6 | 4.5 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.6 | 1.7 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.5 | 10.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.5 | 2.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.5 | 3.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 12.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.4 | 1.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.4 | 2.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.4 | 7.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.4 | 3.8 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.4 | 3.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 1.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 1.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.4 | 5.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 4.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.3 | 4.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.3 | 5.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 1.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.3 | 3.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.5 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.3 | 2.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 1.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 2.6 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.3 | 5.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 13.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 13.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 2.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 7.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.9 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 0.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.2 | 11.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 1.4 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 1.7 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 1.7 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 3.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 1.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 10.3 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.2 | 5.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 1.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 1.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 3.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 4.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 3.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 3.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 4.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 4.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 18.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 3.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 3.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 5.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.4 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 9.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 2.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 1.3 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 2.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 11.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 3.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 36.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.2 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.2 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 10.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 2.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.0 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 2.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 11.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 15.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 2.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 5.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 1.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 10.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 3.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.6 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.1 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 8.5 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 6.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 8.8 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.3 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 24.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 9.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 8.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 4.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 6.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.5 | 6.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.5 | 8.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 2.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 4.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 4.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 7.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 1.8 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 4.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 16.1 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.2 | 3.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 3.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.6 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 5.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 2.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |