Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
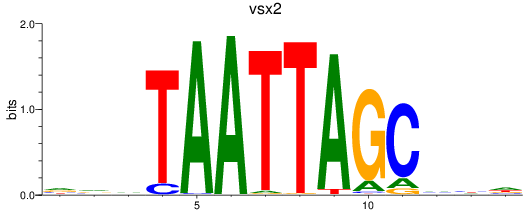
Results for vsx2
Z-value: 1.37
Transcription factors associated with vsx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vsx2
|
ENSDARG00000005574 | visual system homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vsx2 | dr11_v1_chr17_-_31659670_31659670 | 0.84 | 1.8e-26 | Click! |
Activity profile of vsx2 motif
Sorted Z-values of vsx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_21129752 | 31.42 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr23_+_28731379 | 30.69 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr16_+_46111849 | 27.21 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr15_-_16098531 | 26.60 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr20_+_35382482 | 25.85 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr18_-_1185772 | 23.90 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr21_+_28958471 | 18.74 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_-_43356082 | 18.07 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr9_+_11532025 | 17.70 |
ENSDART00000109037
|
cdk5r2b
|
cyclin-dependent kinase 5, regulatory subunit 2b (p39) |
| chr24_-_21923930 | 15.93 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr17_+_31185276 | 15.39 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr1_+_9290103 | 14.74 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr5_-_16996482 | 13.69 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr21_-_27185915 | 13.47 |
ENSDART00000135052
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr15_-_9272328 | 13.04 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr3_-_46817499 | 12.68 |
ENSDART00000013717
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr20_-_9436521 | 12.26 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr23_-_24195519 | 12.20 |
ENSDART00000112370
ENSDART00000180377 |
ano11
|
anoctamin 11 |
| chr4_-_17629444 | 12.09 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr16_+_39159752 | 12.00 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr21_+_25231160 | 11.56 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr13_+_19322686 | 11.24 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr23_+_28582865 | 11.20 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr23_-_24195723 | 10.39 |
ENSDART00000145489
|
ano11
|
anoctamin 11 |
| chr21_+_6751760 | 9.03 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr18_-_2433011 | 8.74 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr18_+_7639401 | 7.93 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr23_+_23658474 | 7.54 |
ENSDART00000162838
|
agrn
|
agrin |
| chr21_+_6751405 | 7.42 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr6_-_40713183 | 7.30 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr24_-_23320223 | 7.26 |
ENSDART00000135846
|
zfhx4
|
zinc finger homeobox 4 |
| chr18_+_28106139 | 7.06 |
ENSDART00000089615
|
kiaa1549lb
|
KIAA1549-like b |
| chr21_+_32820175 | 6.97 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr14_+_25817628 | 6.90 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr23_-_24483311 | 6.81 |
ENSDART00000185793
ENSDART00000109248 |
spen
|
spen family transcriptional repressor |
| chr20_-_46362606 | 6.56 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr7_+_36898850 | 6.54 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr22_-_20011476 | 6.53 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr5_-_64831207 | 6.53 |
ENSDART00000144816
|
lix1
|
limb and CNS expressed 1 |
| chr19_+_22062202 | 6.48 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr15_-_22074315 | 6.39 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr11_+_18873619 | 6.10 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr18_+_14529005 | 5.96 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr12_-_35095414 | 5.53 |
ENSDART00000153229
|
si:dkey-21e13.3
|
si:dkey-21e13.3 |
| chr14_+_21820034 | 5.50 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr6_-_12275836 | 4.92 |
ENSDART00000189980
|
pkp4
|
plakophilin 4 |
| chr12_-_35386910 | 4.85 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr21_-_37790727 | 4.80 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr11_-_37425407 | 4.78 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr11_-_42554290 | 4.76 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr11_+_38280454 | 4.30 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr12_+_30367079 | 4.27 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr20_+_29209615 | 4.16 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr5_-_28109416 | 3.99 |
ENSDART00000042002
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr12_+_30367371 | 3.89 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr17_+_3379673 | 3.85 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr1_+_10318089 | 3.80 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr11_+_18873113 | 3.73 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr14_+_23717165 | 3.62 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr14_+_23874062 | 3.57 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr12_-_19862912 | 3.03 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr20_+_29209767 | 2.95 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_-_46811611 | 2.94 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr7_+_22313533 | 2.73 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr18_-_24989580 | 2.61 |
ENSDART00000163449
|
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr24_-_25004553 | 2.48 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr24_+_39518774 | 2.34 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr21_+_27370671 | 2.30 |
ENSDART00000009234
ENSDART00000142071 |
rbm14a
|
RNA binding motif protein 14a |
| chr22_-_21897203 | 2.21 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr4_+_3980247 | 2.02 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr8_+_18588551 | 2.01 |
ENSDART00000177476
ENSDART00000063539 |
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr20_+_29209926 | 2.01 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr21_+_33172526 | 1.98 |
ENSDART00000183532
|
arl3l1
|
ADP-ribosylation factor-like 3, like 1 |
| chr21_-_32781612 | 1.91 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr24_+_35881124 | 1.81 |
ENSDART00000143015
|
klhl14
|
kelch-like family member 14 |
| chr6_-_32726848 | 1.64 |
ENSDART00000155294
|
zc3h3
|
zinc finger CCCH-type containing 3 |
| chr11_-_29768054 | 1.54 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr13_-_44630111 | 1.52 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr14_-_7207961 | 1.38 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr6_+_28208973 | 1.35 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr19_-_47323267 | 1.25 |
ENSDART00000190077
|
CU138512.1
|
|
| chr22_+_9003090 | 1.22 |
ENSDART00000106414
|
rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr13_+_22295905 | 0.99 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr19_-_19379084 | 0.92 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr20_-_20270191 | 0.84 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr21_-_13661631 | 0.77 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr2_-_26720854 | 0.29 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr23_-_33709964 | 0.20 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr15_+_5360407 | 0.00 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of vsx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.0 | GO:0060074 | synapse maturation(GO:0060074) |
| 2.5 | 7.5 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 2.3 | 6.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 2.3 | 22.6 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 1.4 | 31.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 1.4 | 7.0 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 1.2 | 18.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 1.2 | 11.2 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 1.1 | 13.5 | GO:0042044 | fluid transport(GO:0042044) |
| 1.1 | 26.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.8 | 23.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.5 | 11.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.5 | 6.4 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 | 4.8 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 3.0 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 15.4 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.3 | 2.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 | 4.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.3 | 1.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.3 | 6.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.2 | 6.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 2.0 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.2 | 6.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 6.0 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.2 | 22.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.2 | 13.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 4.9 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 5.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 8.2 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.1 | 1.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 3.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 4.8 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 0.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 4.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 21.0 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.1 | 3.6 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 6.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 13.7 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 12.5 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 2.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 5.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 6.8 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 2.3 | GO:0071773 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 31.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 3.5 | 17.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 3.1 | 18.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.9 | 11.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.8 | 4.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.7 | 11.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.4 | 27.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 12.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 2.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 6.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.2 | 3.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 11.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 4.8 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 13.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 6.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 23.9 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 9.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 8.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 6.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 3.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 9.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 21.4 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 12.5 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 22.1 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 24.5 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 4.9 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 26.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 3.1 | 18.7 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 2.2 | 11.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.4 | 17.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.2 | 7.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.0 | 13.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.8 | 6.9 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.7 | 3.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.7 | 23.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.7 | 31.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.6 | 12.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.6 | 11.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 12.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.5 | 3.8 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.5 | 6.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.3 | 8.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.3 | 7.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.3 | 1.5 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.3 | 27.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.3 | 4.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 2.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 25.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 6.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 4.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 6.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 5.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 9.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.1 | 4.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 2.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 13.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 2.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 9.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 18.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 42.9 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 23.5 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 3.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 14.7 | GO:0022857 | transmembrane transporter activity(GO:0022857) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 17.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 12.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 7.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 18.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.7 | 11.6 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.5 | 7.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 13.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.3 | 6.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |