Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
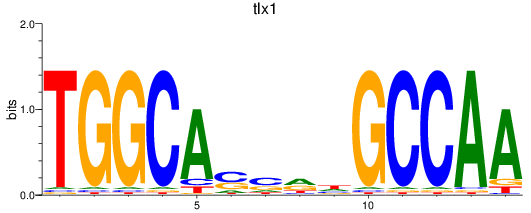
Results for tlx1
Z-value: 1.19
Transcription factors associated with tlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tlx1
|
ENSDARG00000003965 | T cell leukemia homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tlx1 | dr11_v1_chr13_-_28272299_28272299 | -0.10 | 3.6e-01 | Click! |
Activity profile of tlx1 motif
Sorted Z-values of tlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_4146947 | 25.97 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr20_+_10539293 | 19.55 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr20_+_10545514 | 19.16 |
ENSDART00000153667
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr20_+_25581627 | 13.81 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr22_+_38229321 | 12.89 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr12_+_33403694 | 11.08 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr7_+_52122224 | 10.29 |
ENSDART00000174268
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr13_-_9895564 | 9.85 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr3_-_15081874 | 9.41 |
ENSDART00000192532
|
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr25_-_30394746 | 8.88 |
ENSDART00000185346
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr7_+_52135791 | 8.70 |
ENSDART00000098705
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr7_-_52096498 | 7.85 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr14_-_25444774 | 7.53 |
ENSDART00000183448
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr18_+_40462445 | 7.43 |
ENSDART00000087645
|
ugt5c2
|
UDP glucuronosyltransferase 5 family, polypeptide C2 |
| chr13_+_37040789 | 7.16 |
ENSDART00000063412
|
esr2b
|
estrogen receptor 2b |
| chr13_+_28761998 | 6.68 |
ENSDART00000146581
|
prom2
|
prominin 2 |
| chr24_-_21903360 | 6.64 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr11_-_34147205 | 6.53 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr25_-_17378881 | 6.50 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr4_-_14531687 | 6.42 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr7_+_14291323 | 5.70 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr2_-_17827983 | 5.35 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr20_+_34320635 | 5.07 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr7_+_37377335 | 4.81 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr7_-_51953613 | 4.79 |
ENSDART00000142042
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr25_-_31118923 | 4.79 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr15_-_20745513 | 4.56 |
ENSDART00000187438
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr11_+_14937904 | 4.31 |
ENSDART00000185103
|
CR550302.3
|
|
| chr6_-_39199070 | 4.27 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr17_-_50233493 | 3.91 |
ENSDART00000172266
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr8_-_53108207 | 3.89 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr20_+_16750177 | 3.82 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr1_-_56363108 | 3.71 |
ENSDART00000021878
|
gcgrb
|
glucagon receptor b |
| chr4_-_4250317 | 3.68 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr7_+_57109214 | 3.62 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr2_+_30969029 | 3.50 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr4_-_18281070 | 3.29 |
ENSDART00000135330
ENSDART00000179075 ENSDART00000046871 |
uhrf1bp1l
|
UHRF1 binding protein 1-like |
| chr14_+_16036139 | 3.16 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr7_+_57108823 | 3.15 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr3_+_61391636 | 3.11 |
ENSDART00000126417
|
bri3
|
brain protein I3 |
| chr13_+_15682803 | 3.09 |
ENSDART00000188063
|
CR931980.1
|
|
| chr2_-_7800702 | 3.07 |
ENSDART00000146360
|
tbl1xr1b
|
transducin (beta)-like 1 X-linked receptor 1b |
| chr5_+_22791686 | 3.05 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr14_+_31865324 | 3.00 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr2_+_37207461 | 2.96 |
ENSDART00000138952
ENSDART00000132856 ENSDART00000137272 ENSDART00000143468 |
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr8_+_44623540 | 2.95 |
ENSDART00000141513
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr14_+_31865099 | 2.88 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr23_-_38497705 | 2.87 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr6_-_21873266 | 2.84 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr19_-_30403922 | 2.81 |
ENSDART00000181841
|
agr2
|
anterior gradient 2 |
| chr17_+_52822422 | 2.80 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr19_-_30404096 | 2.80 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr23_-_18572521 | 2.79 |
ENSDART00000138599
|
ssr4
|
signal sequence receptor, delta |
| chr7_-_51953807 | 2.72 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr9_-_34296406 | 2.72 |
ENSDART00000125451
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr23_-_18572685 | 2.68 |
ENSDART00000047175
|
ssr4
|
signal sequence receptor, delta |
| chr17_-_31308658 | 2.66 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr8_-_1266181 | 2.62 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr11_+_14284866 | 2.60 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr5_-_57526807 | 2.55 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr20_-_53949798 | 2.41 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr1_+_4101741 | 2.39 |
ENSDART00000163793
|
slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr18_-_2433011 | 2.27 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr7_-_28647959 | 2.22 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr16_+_21426524 | 2.16 |
ENSDART00000182869
|
gsdmeb
|
gasdermin Eb |
| chr11_+_37265692 | 2.15 |
ENSDART00000184691
|
il17rc
|
interleukin 17 receptor C |
| chr19_-_8926575 | 2.13 |
ENSDART00000080897
|
rprd2a
|
regulation of nuclear pre-mRNA domain containing 2a |
| chr18_-_127873 | 2.03 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr1_-_58059134 | 1.99 |
ENSDART00000160970
|
caspb
|
caspase b |
| chr18_-_39473055 | 1.98 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr23_-_36446307 | 1.94 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr17_+_10578823 | 1.93 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr16_-_22585289 | 1.91 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr18_+_7543347 | 1.82 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr23_+_21278948 | 1.75 |
ENSDART00000156701
ENSDART00000033970 |
ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr5_+_31283576 | 1.73 |
ENSDART00000133743
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr18_+_41560822 | 1.69 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr2_-_59285085 | 1.60 |
ENSDART00000131880
|
ftr34
|
finTRIM family, member 34 |
| chr7_+_47316944 | 1.60 |
ENSDART00000173534
|
dpy19l3
|
dpy-19 like C-mannosyltransferase 3 |
| chr3_-_34599662 | 1.58 |
ENSDART00000055259
|
nmrk1
|
nicotinamide riboside kinase 1 |
| chr22_-_22416337 | 1.46 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr6_+_2951645 | 1.46 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr11_-_103136 | 1.46 |
ENSDART00000173308
ENSDART00000162982 |
elmo2
|
engulfment and cell motility 2 |
| chr8_+_44722140 | 1.45 |
ENSDART00000163381
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr8_-_31053872 | 1.43 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr3_-_37681824 | 1.40 |
ENSDART00000185858
|
gpatch8
|
G patch domain containing 8 |
| chr9_-_24242592 | 1.36 |
ENSDART00000039399
|
cavin2a
|
caveolae associated protein 2a |
| chr3_+_24482999 | 1.36 |
ENSDART00000059179
|
nptxra
|
neuronal pentraxin receptor a |
| chr3_+_57038033 | 1.29 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr15_-_38202630 | 1.25 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr20_-_3390406 | 1.25 |
ENSDART00000136987
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr8_+_9699111 | 1.24 |
ENSDART00000111853
|
gripap1
|
GRIP1 associated protein 1 |
| chr24_+_17262879 | 1.23 |
ENSDART00000145949
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr22_-_16400484 | 1.18 |
ENSDART00000135987
|
lama3
|
laminin, alpha 3 |
| chr8_-_39822917 | 1.18 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr19_+_40379771 | 1.14 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr24_+_7884880 | 1.10 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr2_+_50722439 | 1.02 |
ENSDART00000188927
|
fyco1b
|
FYVE and coiled-coil domain containing 1b |
| chr16_+_14029283 | 0.99 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr13_-_35760969 | 0.95 |
ENSDART00000127476
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr16_-_9453591 | 0.94 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr13_-_15865335 | 0.94 |
ENSDART00000135186
|
si:ch211-57f7.7
|
si:ch211-57f7.7 |
| chr5_-_30074332 | 0.93 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr11_-_13341483 | 0.91 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr16_-_11928774 | 0.87 |
ENSDART00000183229
|
cd4-1
|
CD4-1 molecule |
| chr15_-_18091157 | 0.85 |
ENSDART00000153848
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr23_-_45504991 | 0.85 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr5_+_28398449 | 0.85 |
ENSDART00000165292
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr13_+_18371208 | 0.84 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr5_-_32338866 | 0.83 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr13_-_35761266 | 0.82 |
ENSDART00000190217
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr11_-_13341051 | 0.78 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr2_-_59285407 | 0.74 |
ENSDART00000181616
|
ftr34
|
finTRIM family, member 34 |
| chr14_-_32503363 | 0.72 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr2_-_59303338 | 0.72 |
ENSDART00000100987
ENSDART00000140840 |
ftr35
|
finTRIM family, member 35 |
| chr23_-_6765653 | 0.71 |
ENSDART00000192310
|
FP102169.1
|
|
| chr6_+_53349966 | 0.70 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr22_+_11857356 | 0.70 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr7_-_24472991 | 0.69 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr5_+_63329608 | 0.69 |
ENSDART00000139180
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr7_-_22132265 | 0.65 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr17_-_22067451 | 0.63 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr12_-_2518178 | 0.63 |
ENSDART00000128677
ENSDART00000086510 |
mapk8b
|
mitogen-activated protein kinase 8b |
| chr2_-_37352514 | 0.58 |
ENSDART00000140498
ENSDART00000186422 |
skila
|
SKI-like proto-oncogene a |
| chr17_+_19630068 | 0.56 |
ENSDART00000182619
|
rgs7a
|
regulator of G protein signaling 7a |
| chr14_+_21820034 | 0.55 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr15_+_23528010 | 0.53 |
ENSDART00000152786
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr1_+_49568335 | 0.53 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr11_-_17755444 | 0.52 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr20_-_29499363 | 0.51 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr4_+_74131530 | 0.46 |
ENSDART00000174125
|
CABZ01072523.1
|
|
| chr9_+_16449398 | 0.45 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr10_-_29827000 | 0.40 |
ENSDART00000131418
|
zpr1
|
ZPR1 zinc finger |
| chr12_+_13405445 | 0.38 |
ENSDART00000089042
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr5_+_61839707 | 0.36 |
ENSDART00000163051
|
CR759879.1
|
Danio rerio interferon-induced protein with tetratricopeptide repeats 5 (LOC572297), mRNA. |
| chr19_-_47571797 | 0.35 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr9_-_25089536 | 0.34 |
ENSDART00000143479
|
rubcnl
|
RUN and cysteine rich domain containing beclin 1 interacting protein like |
| chr8_-_16712111 | 0.26 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr3_-_19561058 | 0.26 |
ENSDART00000079323
|
zgc:163079
|
zgc:163079 |
| chr21_+_39336285 | 0.22 |
ENSDART00000139677
|
si:ch211-274p24.4
|
si:ch211-274p24.4 |
| chr8_-_34052019 | 0.19 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr3_-_27061637 | 0.19 |
ENSDART00000157126
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr9_-_44939104 | 0.18 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr10_-_7857494 | 0.17 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr1_+_35194454 | 0.16 |
ENSDART00000165389
|
sc:d189
|
sc:d189 |
| chr6_+_39184236 | 0.10 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr15_+_22722684 | 0.07 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr4_-_46648979 | 0.06 |
ENSDART00000162101
|
BX927193.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of tlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 9.8 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 1.1 | 54.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.8 | 6.8 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.7 | 7.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.7 | 5.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.7 | 2.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.6 | 51.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.6 | 9.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.6 | 4.6 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.6 | 5.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.5 | 1.6 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.5 | 26.0 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.4 | 1.2 | GO:0097065 | anterior head development(GO:0097065) |
| 0.3 | 4.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 | 1.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.3 | 7.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.3 | 2.6 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 1.4 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.2 | 2.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 0.5 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 3.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 1.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 5.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 2.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.8 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.9 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 3.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 3.0 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 1.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 3.7 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 3.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.3 | GO:0016117 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 2.0 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 1.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.6 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 3.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 6.8 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.1 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 3.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 3.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 1.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.9 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 9.9 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 2.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 2.7 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 1.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 1.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 2.2 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 1.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 5.3 | GO:0072657 | protein localization to membrane(GO:0072657) |
| 0.0 | 4.7 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 3.9 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.7 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 1.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.5 | GO:0006909 | phagocytosis(GO:0006909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.6 | 3.0 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.5 | 2.0 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 1.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 0.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 0.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 1.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 2.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.1 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 4.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 0.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 40.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 1.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 3.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 13.4 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 3.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 30.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 11.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 3.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.2 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.8 | 54.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.8 | 9.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.7 | 3.7 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.6 | 4.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.5 | 3.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.4 | 2.8 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.4 | 7.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.3 | 38.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 9.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 12.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 1.7 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 5.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 3.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 2.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 2.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 2.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 5.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 7.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.9 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 6.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 3.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.9 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 2.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 6.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 3.9 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.3 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 5.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 7.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.2 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0034594 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 7.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 16.5 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 6.5 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 3.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 21.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 4.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 11.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 3.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 6.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 26.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.9 | 11.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.8 | 4.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.5 | 10.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 21.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 3.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 9.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 5.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |