Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
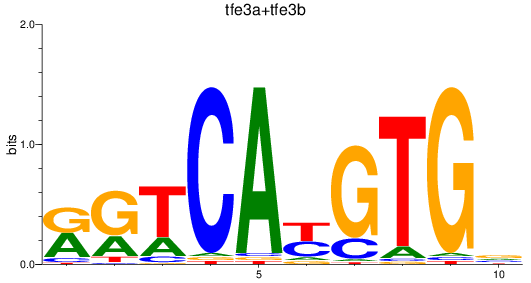
Results for tfe3a+tfe3b
Z-value: 1.49
Transcription factors associated with tfe3a+tfe3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfe3b
|
ENSDARG00000019457 | transcription factor binding to IGHM enhancer 3b |
|
tfe3a
|
ENSDARG00000098903 | transcription factor binding to IGHM enhancer 3a |
|
tfe3b
|
ENSDARG00000111231 | transcription factor binding to IGHM enhancer 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfe3b | dr11_v1_chr11_+_25504215_25504215 | 0.37 | 2.1e-04 | Click! |
| tfe3a | dr11_v1_chr8_+_7778770_7778770 | -0.15 | 1.4e-01 | Click! |
Activity profile of tfe3a+tfe3b motif
Sorted Z-values of tfe3a+tfe3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_31665836 | 23.14 |
ENSDART00000135411
ENSDART00000143914 |
si:ch211-106h4.12
|
si:ch211-106h4.12 |
| chr16_+_23960933 | 22.55 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr5_-_29534748 | 15.49 |
ENSDART00000159587
|
AL831768.1
|
|
| chr16_-_45178430 | 15.07 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr3_-_16784280 | 14.86 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr14_+_52571134 | 13.49 |
ENSDART00000166708
|
rpl26
|
ribosomal protein L26 |
| chr18_-_48517040 | 12.00 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr22_-_17677947 | 11.70 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr20_-_17041025 | 11.66 |
ENSDART00000063764
|
si:dkey-5n18.1
|
si:dkey-5n18.1 |
| chr16_+_23960744 | 11.35 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr18_-_7539166 | 11.32 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr25_+_37443194 | 10.63 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr21_-_25741096 | 10.15 |
ENSDART00000181756
|
cldnh
|
claudin h |
| chr18_+_44703343 | 10.10 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr20_+_38257766 | 10.07 |
ENSDART00000147485
ENSDART00000149160 |
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr2_-_24069331 | 9.74 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr10_+_36662640 | 9.58 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr6_+_112579 | 9.49 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr12_-_3903886 | 9.34 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr3_-_39488639 | 9.32 |
ENSDART00000161644
|
zgc:100868
|
zgc:100868 |
| chr18_-_1414760 | 9.13 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr3_-_39488482 | 9.06 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr23_-_5719453 | 9.04 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr13_-_214122 | 8.92 |
ENSDART00000169273
|
ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr16_+_23961276 | 8.80 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr3_+_42923275 | 8.66 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr25_+_245438 | 8.40 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr25_+_10547228 | 8.37 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr9_-_56272465 | 8.34 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr14_-_15171435 | 8.30 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr19_+_791538 | 8.30 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr1_+_54766943 | 8.20 |
ENSDART00000144759
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr3_+_16922226 | 8.05 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr17_+_25331576 | 8.01 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr13_-_36050303 | 8.01 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr10_-_35149513 | 8.01 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr19_+_19762183 | 8.00 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr1_+_14253118 | 7.99 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr19_+_19772765 | 7.99 |
ENSDART00000182028
ENSDART00000161019 |
hoxa3a
|
homeobox A3a |
| chr7_+_49654588 | 7.98 |
ENSDART00000025451
ENSDART00000141934 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr5_+_27404946 | 7.96 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr15_-_1198886 | 7.81 |
ENSDART00000063285
|
lxn
|
latexin |
| chr2_-_43545342 | 7.77 |
ENSDART00000179796
|
CABZ01058721.1
|
|
| chr1_-_58059134 | 7.63 |
ENSDART00000160970
|
caspb
|
caspase b |
| chr19_+_48117995 | 7.62 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr11_+_24729346 | 7.50 |
ENSDART00000087740
|
zgc:153953
|
zgc:153953 |
| chr7_-_19923249 | 7.50 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr6_-_40429411 | 7.34 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr8_+_30797363 | 7.28 |
ENSDART00000077280
|
mmp11a
|
matrix metallopeptidase 11a |
| chr11_+_10541258 | 7.27 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr24_-_42072886 | 7.16 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr7_+_39410180 | 7.12 |
ENSDART00000168641
|
CT030188.1
|
|
| chr5_+_26204561 | 7.09 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr7_-_55633475 | 7.02 |
ENSDART00000149478
|
galns
|
galactosamine (N-acetyl)-6-sulfatase |
| chr17_-_24916889 | 6.99 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr9_-_23944470 | 6.99 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr9_+_35077546 | 6.97 |
ENSDART00000142688
|
si:dkey-192g7.3
|
si:dkey-192g7.3 |
| chr3_+_31662126 | 6.96 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr24_+_42149453 | 6.90 |
ENSDART00000128766
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr23_+_384850 | 6.84 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr18_-_6943577 | 6.84 |
ENSDART00000132399
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr21_-_22673758 | 6.75 |
ENSDART00000164910
|
gig2i
|
grass carp reovirus (GCRV)-induced gene 2i |
| chr11_-_270210 | 6.73 |
ENSDART00000005217
ENSDART00000172779 |
alas1
|
aminolevulinate, delta-, synthase 1 |
| chr10_+_41159241 | 6.65 |
ENSDART00000141657
|
anxa4
|
annexin A4 |
| chr11_-_42730063 | 6.63 |
ENSDART00000169776
|
si:ch73-106k19.2
|
si:ch73-106k19.2 |
| chr20_-_32045057 | 6.58 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr17_+_30843881 | 6.57 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr19_-_1947403 | 6.54 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr24_-_12938922 | 6.51 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr25_+_19718504 | 6.50 |
ENSDART00000036702
|
mxd
|
myxovirus (influenza virus) resistance D |
| chr17_+_6563307 | 6.50 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr12_-_44180132 | 6.48 |
ENSDART00000165998
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr16_+_19029297 | 6.45 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr23_+_43201142 | 6.41 |
ENSDART00000158832
ENSDART00000190315 |
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr23_+_43200911 | 6.39 |
ENSDART00000164262
|
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr17_+_32622933 | 6.38 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr18_+_26422124 | 6.29 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr2_+_36007449 | 6.21 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr8_+_25615781 | 6.17 |
ENSDART00000062385
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr1_+_12135129 | 6.16 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr9_-_34269066 | 6.16 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr3_-_3209432 | 6.15 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr15_+_20403903 | 6.13 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr15_-_43625549 | 6.12 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr19_+_2590182 | 6.10 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr2_+_24507770 | 6.04 |
ENSDART00000154802
ENSDART00000052063 |
rps28
|
ribosomal protein S28 |
| chr24_+_35564668 | 6.04 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr7_-_67214972 | 6.00 |
ENSDART00000156861
|
swap70a
|
switching B cell complex subunit SWAP70a |
| chr4_-_992063 | 5.97 |
ENSDART00000181630
ENSDART00000183898 ENSDART00000160902 |
naga
|
N-acetylgalactosaminidase, alpha |
| chr10_-_24724388 | 5.97 |
ENSDART00000148582
|
smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr18_+_44649804 | 5.92 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr7_+_39410393 | 5.87 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr10_+_36650222 | 5.87 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr23_+_26079467 | 5.87 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr18_-_7539469 | 5.85 |
ENSDART00000101296
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr5_+_4564233 | 5.82 |
ENSDART00000193435
|
CABZ01058647.1
|
|
| chr8_-_31075015 | 5.76 |
ENSDART00000010993
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr9_-_33063083 | 5.74 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr15_-_47468085 | 5.71 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr5_-_1963498 | 5.58 |
ENSDART00000073462
|
rplp0
|
ribosomal protein, large, P0 |
| chr14_+_22076596 | 5.57 |
ENSDART00000106147
ENSDART00000100278 ENSDART00000131489 |
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr19_-_808265 | 5.56 |
ENSDART00000082454
|
glmp
|
glycosylated lysosomal membrane protein |
| chr22_+_29991834 | 5.52 |
ENSDART00000147728
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr18_-_48550426 | 5.46 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr22_+_5120033 | 5.45 |
ENSDART00000169200
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr19_-_15229421 | 5.28 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr3_+_14463941 | 5.12 |
ENSDART00000170927
|
cnn1b
|
calponin 1, basic, smooth muscle, b |
| chr15_+_46853252 | 5.12 |
ENSDART00000186040
|
zgc:153039
|
zgc:153039 |
| chr23_+_45910033 | 5.11 |
ENSDART00000165025
|
abcg2a
|
ATP-binding cassette, sub-family G (WHITE), member 2a |
| chr19_-_42503143 | 5.07 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr7_-_59054322 | 5.05 |
ENSDART00000165390
|
chmp5b
|
charged multivesicular body protein 5b |
| chr15_-_4580763 | 5.04 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr6_+_392815 | 4.99 |
ENSDART00000163142
|
cyth4b
|
cytohesin 4b |
| chr4_+_60664235 | 4.98 |
ENSDART00000186182
|
CABZ01016118.1
|
|
| chr8_+_13389115 | 4.89 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr1_+_58176023 | 4.88 |
ENSDART00000151051
|
si:ch211-15j1.7
|
si:ch211-15j1.7 |
| chr19_+_19786117 | 4.86 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr7_-_3693303 | 4.77 |
ENSDART00000166949
|
si:ch211-282j17.13
|
si:ch211-282j17.13 |
| chr23_+_35847200 | 4.77 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
| chr20_-_39735952 | 4.72 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr12_+_20336070 | 4.72 |
ENSDART00000066385
|
zgc:163057
|
zgc:163057 |
| chr6_-_40651944 | 4.70 |
ENSDART00000187423
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr5_+_50913034 | 4.69 |
ENSDART00000149787
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr10_-_15053507 | 4.69 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr25_+_22107643 | 4.68 |
ENSDART00000089680
|
sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr21_-_10773344 | 4.68 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr15_-_43327911 | 4.66 |
ENSDART00000077386
|
prss16
|
protease, serine, 16 (thymus) |
| chr21_+_4540127 | 4.64 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr17_+_32623931 | 4.63 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr18_-_46280578 | 4.62 |
ENSDART00000131724
|
pld3
|
phospholipase D family, member 3 |
| chr25_+_258883 | 4.60 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr1_-_8140763 | 4.60 |
ENSDART00000091508
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr15_-_3277635 | 4.60 |
ENSDART00000189094
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr4_+_5341592 | 4.60 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr10_+_5268054 | 4.60 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr21_+_7298687 | 4.60 |
ENSDART00000187746
|
CU929160.1
|
|
| chr7_-_35126374 | 4.59 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr2_-_10386738 | 4.58 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr7_-_11383933 | 4.57 |
ENSDART00000163949
|
mesd
|
mesoderm development LRP chaperone |
| chr12_+_37401331 | 4.57 |
ENSDART00000125040
|
si:ch211-152f22.4
|
si:ch211-152f22.4 |
| chr23_+_39089574 | 4.56 |
ENSDART00000164711
|
nfatc2a
|
nuclear factor of activated T cells 2a |
| chr19_+_19756425 | 4.52 |
ENSDART00000167606
|
hoxa3a
|
homeobox A3a |
| chr5_-_23800376 | 4.52 |
ENSDART00000134184
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr10_-_21953643 | 4.51 |
ENSDART00000188921
ENSDART00000193569 |
FO744833.2
|
|
| chr3_-_18805225 | 4.51 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr16_+_33143503 | 4.49 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr5_+_50913357 | 4.45 |
ENSDART00000092938
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr19_-_1855368 | 4.43 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr22_-_17653143 | 4.41 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr25_+_31323978 | 4.36 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr7_-_3669868 | 4.34 |
ENSDART00000172907
|
si:ch211-282j17.12
|
si:ch211-282j17.12 |
| chr14_+_39156 | 4.34 |
ENSDART00000082184
|
tmem107
|
transmembrane protein 107 |
| chr15_-_28596507 | 4.31 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr21_-_22715297 | 4.29 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr8_-_16650595 | 4.29 |
ENSDART00000135319
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr2_+_24507597 | 4.29 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr25_+_20188769 | 4.28 |
ENSDART00000142781
|
cald1b
|
caldesmon 1b |
| chr5_+_51594209 | 4.25 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr8_+_41229233 | 4.25 |
ENSDART00000131135
|
zgc:152830
|
zgc:152830 |
| chr19_+_47502581 | 4.24 |
ENSDART00000171526
|
CU695215.2
|
|
| chr11_+_45092866 | 4.21 |
ENSDART00000163408
|
si:dkey-93h22.8
|
si:dkey-93h22.8 |
| chr23_-_26784736 | 4.21 |
ENSDART00000024064
ENSDART00000131615 |
galnt6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr6_+_7123980 | 4.20 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr24_-_18919562 | 4.12 |
ENSDART00000144244
ENSDART00000106188 ENSDART00000182518 |
cpa6
|
carboxypeptidase A6 |
| chr4_-_75930031 | 4.12 |
ENSDART00000170728
ENSDART00000181924 |
si:dkey-261j11.3
si:dkey-261j11.2
|
si:dkey-261j11.3 si:dkey-261j11.2 |
| chr22_-_10165446 | 4.06 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr6_-_2154137 | 4.05 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr21_-_3007412 | 4.04 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr5_-_69944084 | 4.03 |
ENSDART00000188557
ENSDART00000127782 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr9_+_55857193 | 4.02 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr23_+_10146542 | 4.01 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr18_-_11729 | 4.00 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr19_+_34169055 | 3.96 |
ENSDART00000135592
ENSDART00000186043 |
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr8_+_8671229 | 3.96 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr13_-_280652 | 3.96 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr21_-_3700334 | 3.94 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr25_+_29474982 | 3.94 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr11_-_42472941 | 3.92 |
ENSDART00000166624
|
arf4b
|
ADP-ribosylation factor 4b |
| chr23_+_13814978 | 3.90 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr7_-_11384279 | 3.89 |
ENSDART00000102515
ENSDART00000172796 |
mesd
|
mesoderm development LRP chaperone |
| chr14_+_23709543 | 3.89 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr21_-_22669669 | 3.76 |
ENSDART00000101784
|
gig2j
|
grass carp reovirus (GCRV)-induced gene 2j |
| chr3_-_4591643 | 3.73 |
ENSDART00000138144
|
ftr50
|
finTRIM family, member 50 |
| chr20_-_37813863 | 3.72 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr15_+_20403084 | 3.71 |
ENSDART00000141388
ENSDART00000152734 |
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr15_+_1199407 | 3.67 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr9_+_37152564 | 3.65 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr6_+_7490154 | 3.63 |
ENSDART00000014892
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr8_+_21195420 | 3.62 |
ENSDART00000100234
ENSDART00000091307 |
col2a1a
|
collagen, type II, alpha 1a |
| chr8_+_20438884 | 3.61 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr3_-_8765165 | 3.60 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr1_-_59139599 | 3.58 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr13_+_7049210 | 3.58 |
ENSDART00000168608
ENSDART00000128675 ENSDART00000164789 |
rnaset2
|
ribonuclease T2 |
| chr24_+_7631797 | 3.58 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr14_+_16765992 | 3.58 |
ENSDART00000140061
|
sqstm1
|
sequestosome 1 |
| chr23_+_35847538 | 3.56 |
ENSDART00000143935
|
rarga
|
retinoic acid receptor gamma a |
| chr13_+_22717366 | 3.53 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr21_-_7265219 | 3.51 |
ENSDART00000158852
|
egfl7
|
EGF-like-domain, multiple 7 |
| chr24_+_27511734 | 3.50 |
ENSDART00000105771
|
cxcl32b.1
|
chemokine (C-X-C motif) ligand 32b, duplicate 1 |
| chr21_-_45891262 | 3.50 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfe3a+tfe3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.2 | 42.7 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 3.3 | 23.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 2.7 | 8.0 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 2.0 | 8.0 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 2.0 | 8.0 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 1.7 | 8.5 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 1.6 | 9.8 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 1.6 | 4.9 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 1.6 | 6.5 | GO:0071548 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 1.5 | 6.2 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 1.5 | 4.6 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 1.5 | 12.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 1.5 | 6.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 1.2 | 11.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 1.1 | 3.4 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 1.0 | 2.9 | GO:0035973 | aggrephagy(GO:0035973) |
| 1.0 | 2.9 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.9 | 4.6 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.9 | 3.6 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.9 | 2.7 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.9 | 2.7 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.9 | 2.6 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.9 | 6.0 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.8 | 3.4 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.8 | 5.7 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.8 | 4.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.8 | 2.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.8 | 2.3 | GO:1902102 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.8 | 10.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.8 | 5.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.7 | 7.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.7 | 15.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.7 | 5.0 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.7 | 2.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.7 | 3.5 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.7 | 2.7 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.7 | 4.6 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.6 | 3.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.6 | 8.1 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.6 | 16.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.6 | 1.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.5 | 3.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.5 | 1.6 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.5 | 5.6 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.5 | 0.5 | GO:0048753 | pigment granule organization(GO:0048753) |
| 0.5 | 7.6 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.5 | 7.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.5 | 14.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.4 | 10.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 3.6 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.4 | 3.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 4.5 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.4 | 4.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.4 | 6.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.4 | 8.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.4 | 1.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.4 | 8.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 8.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 3.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.3 | 6.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.3 | 6.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.3 | 3.4 | GO:0090148 | membrane fission(GO:0090148) |
| 0.3 | 3.6 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 4.6 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.3 | 4.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 0.9 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.3 | 2.5 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.3 | 2.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 3.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 13.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.3 | 4.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 4.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 2.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.3 | 7.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.3 | 0.9 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.3 | 1.7 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.3 | 2.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.3 | 3.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.3 | 4.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.3 | 1.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.3 | 1.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 14.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.9 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 1.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 7.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.2 | 4.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.2 | 2.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.2 | 2.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 1.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 0.7 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.2 | 0.7 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.2 | 1.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.2 | 4.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 3.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 3.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.2 | 3.1 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.2 | 2.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.3 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 4.7 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.2 | 6.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.2 | 7.5 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.2 | 2.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 3.1 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 1.6 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 4.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 4.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.2 | 0.6 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 1.9 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.2 | 6.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 1.3 | GO:0002312 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.2 | 2.6 | GO:0060021 | palate development(GO:0060021) |
| 0.2 | 1.1 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.2 | 2.4 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 2.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 2.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 2.6 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.2 | 3.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.2 | 2.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 2.8 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 2.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 5.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 1.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 1.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.2 | 3.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 6.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 1.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 3.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 10.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 9.9 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.1 | 9.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 5.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 2.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 1.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 2.5 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 1.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.4 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.1 | 7.0 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 5.6 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 1.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 1.8 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 2.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 1.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 2.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 3.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 6.1 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 4.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.4 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 8.0 | GO:0051170 | nuclear import(GO:0051170) |
| 0.1 | 3.0 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 8.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 0.4 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 2.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 7.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 2.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 7.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 1.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 12.2 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.1 | 6.1 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 3.5 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.5 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 1.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 1.0 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.2 | GO:0060623 | regulation of sister chromatid cohesion(GO:0007063) regulation of chromosome condensation(GO:0060623) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 | 2.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 2.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 6.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.4 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 2.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 2.0 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 2.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 2.0 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 1.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 2.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.0 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.8 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 1.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 | 4.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.8 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 0.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 4.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 2.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 1.9 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 1.3 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 1.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.2 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 2.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.6 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.3 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 2.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 1.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 2.6 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 26.6 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 1.4 | GO:0022406 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 2.1 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.7 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.1 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.7 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 1.2 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 2.8 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 2.0 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.0 | 0.6 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.1 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0007168 | cGMP biosynthetic process(GO:0006182) receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.9 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.4 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.9 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.4 | GO:0051225 | spindle assembly(GO:0051225) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 42.7 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 3.4 | 10.3 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 1.9 | 7.6 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.7 | 5.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.2 | 3.6 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 1.2 | 4.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.0 | 5.9 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.9 | 3.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.8 | 3.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.7 | 8.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.7 | 2.8 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.7 | 2.6 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.6 | 2.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.5 | 3.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.5 | 9.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 1.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 3.5 | GO:0030428 | cell septum(GO:0030428) |
| 0.4 | 1.7 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.4 | 2.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 4.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 2.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.4 | 2.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.4 | 9.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.4 | 2.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.3 | 2.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 6.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 3.0 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 0.9 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.3 | 5.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 4.6 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 5.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 4.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.3 | 2.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 1.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.2 | 11.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 64.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.2 | 1.0 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.2 | 2.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 2.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 3.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 4.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 11.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 6.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 1.5 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 23.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 0.8 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.2 | 4.0 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 8.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 3.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 10.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 9.6 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 1.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 2.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 10.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 4.8 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.7 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 2.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 4.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 3.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 2.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 3.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 2.8 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 4.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 0.8 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 4.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 2.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 16.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 6.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 42.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.4 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 12.8 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 5.0 | GO:0043292 | myofibril(GO:0030016) contractile fiber(GO:0043292) |
| 0.0 | 0.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 2.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 7.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 10.8 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.7 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.2 | 42.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 3.9 | 15.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 2.4 | 7.3 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 2.3 | 6.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 2.2 | 6.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 2.2 | 6.6 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 2.0 | 8.0 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 2.0 | 6.0 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 1.8 | 10.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.3 | 5.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.2 | 17.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.2 | 6.0 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 1.1 | 5.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 1.0 | 4.0 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.9 | 2.8 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.9 | 2.8 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.9 | 2.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.9 | 5.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.9 | 3.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.9 | 2.7 | GO:0042806 | fucose binding(GO:0042806) |
| 0.9 | 3.5 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.8 | 3.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.8 | 5.5 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.8 | 2.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.7 | 3.6 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.7 | 2.2 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.7 | 2.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.6 | 2.6 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.6 | 16.0 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.6 | 7.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.6 | 3.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) galactosidase activity(GO:0015925) |
| 0.6 | 4.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.6 | 7.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 7.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 7.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.6 | 3.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.6 | 3.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 2.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.5 | 4.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.5 | 1.5 | GO:0034618 | arginine binding(GO:0034618) |
| 0.5 | 6.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 12.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 2.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 9.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.4 | 3.5 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.4 | 3.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 9.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 2.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 4.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 4.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 2.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.4 | 1.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 2.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 3.6 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 3.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 4.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.3 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.3 | 4.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 3.4 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 4.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 0.9 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.3 | 13.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 10.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.3 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 1.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 7.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 4.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 3.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 4.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 1.0 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 1.0 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 1.4 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.2 | 1.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 0.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.2 | 3.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 4.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 2.4 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 3.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 4.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.2 | 31.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 3.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 3.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 3.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 3.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 4.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 5.6 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 0.9 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 4.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 5.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.2 | 2.4 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.2 | 1.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.2 | 4.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 3.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.2 | 4.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 15.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 6.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.5 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.3 | GO:0008296 | deoxyribonuclease I activity(GO:0004530) 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 2.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 27.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 3.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.7 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 3.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 6.1 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 5.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 7.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 0.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 4.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 14.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 5.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 2.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 3.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 5.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 2.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.5 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 4.7 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 11.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 1.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 3.5 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 10.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 4.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 4.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 4.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 1.1 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 12.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 4.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 17.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.2 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 2.5 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.0 | 3.8 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.9 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 3.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 5.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 2.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 3.3 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.5 | 23.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.5 | 6.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 4.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 4.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 3.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.3 | 3.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 14.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.3 | 6.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 4.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 7.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 10.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 4.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 18.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 2.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.5 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.7 | 8.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.6 | 8.4 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.5 | 7.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.5 | 5.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.5 | 7.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.4 | 4.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.4 | 4.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.4 | 4.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 12.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 34.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 6.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.3 | 11.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.3 | 15.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 15.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.2 | 1.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 5.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 3.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.2 | 7.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 2.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 7.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 10.9 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 6.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 8.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.2 | 4.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 6.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 3.0 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 5.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 5.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.8 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 2.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 6.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 5.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 2.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |