Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
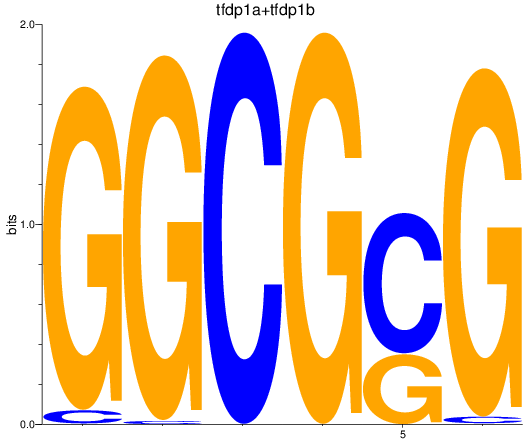
Results for tfdp1a+tfdp1b
Z-value: 3.20
Transcription factors associated with tfdp1a+tfdp1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfdp1b
|
ENSDARG00000016304 | transcription factor Dp-1, b |
|
tfdp1a
|
ENSDARG00000019293 | transcription factor Dp-1, a |
|
tfdp1a
|
ENSDARG00000111589 | transcription factor Dp-1, a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfdp1a | dr11_v1_chr9_+_34950942_34950942 | 0.64 | 2.8e-12 | Click! |
| tfdp1b | dr11_v1_chr1_+_230363_230363 | 0.28 | 6.2e-03 | Click! |
Activity profile of tfdp1a+tfdp1b motif
Sorted Z-values of tfdp1a+tfdp1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_41305748 | 45.34 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr4_+_5741733 | 41.77 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr9_-_44295071 | 34.14 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr18_+_18104235 | 31.52 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr22_-_11493236 | 29.39 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr19_+_31771270 | 28.85 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr4_-_5764255 | 28.62 |
ENSDART00000113864
|
faxca
|
failed axon connections homolog a |
| chr14_+_35748385 | 27.92 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr14_+_35748206 | 27.64 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr18_-_5692292 | 27.33 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr13_+_33117528 | 26.68 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr19_-_27966526 | 26.59 |
ENSDART00000141896
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr15_+_4632782 | 26.38 |
ENSDART00000156012
|
si:dkey-35i13.1
|
si:dkey-35i13.1 |
| chr3_-_59472422 | 26.33 |
ENSDART00000100327
|
nptx1l
|
neuronal pentraxin 1 like |
| chr10_-_15405564 | 26.22 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr2_+_42724404 | 26.01 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr16_-_12953739 | 24.16 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr23_-_35694171 | 24.04 |
ENSDART00000077539
|
tuba1c
|
tubulin, alpha 1c |
| chr19_+_23982466 | 23.80 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr16_-_43025885 | 23.51 |
ENSDART00000193146
ENSDART00000157302 |
si:dkey-7j14.5
|
si:dkey-7j14.5 |
| chr14_-_2163454 | 23.21 |
ENSDART00000160123
ENSDART00000169653 |
pcdh2ab9
pcdh2ac
|
protocadherin 2 alpha b 9 protocadherin 2 alpha c |
| chr3_+_31933893 | 23.18 |
ENSDART00000146509
ENSDART00000139644 |
lin7b
|
lin-7 homolog B (C. elegans) |
| chr19_-_9472893 | 22.71 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr10_-_22845485 | 22.46 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr4_-_18201622 | 22.19 |
ENSDART00000133509
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr1_-_50859053 | 21.84 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr16_+_32059785 | 21.65 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr8_+_31821396 | 21.14 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr11_-_3552067 | 21.01 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr13_+_27314795 | 20.90 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr11_+_7324704 | 20.69 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr16_-_12173554 | 20.61 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr21_-_28340977 | 20.31 |
ENSDART00000141629
|
nrxn2a
|
neurexin 2a |
| chr1_-_21409877 | 19.94 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr3_-_30123113 | 19.31 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr18_-_8312848 | 18.96 |
ENSDART00000092033
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr17_-_17447899 | 18.78 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr22_-_21150845 | 18.37 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr1_+_14348399 | 18.29 |
ENSDART00000064468
|
crmp1
|
collapsin response mediator protein 1 |
| chr6_+_36942966 | 17.87 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr14_-_30642819 | 17.72 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr2_+_31833997 | 17.44 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr21_+_6780340 | 17.38 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr6_+_34512313 | 17.20 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr19_+_37857936 | 17.13 |
ENSDART00000189289
|
nxph1
|
neurexophilin 1 |
| chr23_+_40461411 | 16.79 |
ENSDART00000184223
|
soga3b
|
SOGA family member 3b |
| chr24_+_24014880 | 16.75 |
ENSDART00000041335
|
chodl
|
chondrolectin |
| chr19_-_10243148 | 16.42 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr10_+_21730585 | 16.38 |
ENSDART00000188576
|
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr19_-_27966780 | 16.33 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr13_+_421231 | 16.20 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr7_-_28148310 | 16.13 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr5_+_23118470 | 16.06 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr21_+_5169154 | 16.03 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr21_-_34658266 | 15.93 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr21_+_11468934 | 15.86 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr16_+_46111849 | 15.72 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr14_+_5117072 | 15.68 |
ENSDART00000189628
|
nanos1
|
nanos homolog 1 |
| chr21_+_31150438 | 15.45 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_+_29798064 | 15.26 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr20_+_18209895 | 15.26 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr4_-_9810999 | 15.15 |
ENSDART00000146858
|
si:dkeyp-27e10.3
|
si:dkeyp-27e10.3 |
| chr13_+_12299997 | 15.06 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr1_-_14233815 | 15.03 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr18_-_44610992 | 14.93 |
ENSDART00000125968
ENSDART00000185836 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr5_+_4054704 | 14.84 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr9_-_48214216 | 14.83 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr8_-_40327397 | 14.81 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr2_-_43605556 | 14.67 |
ENSDART00000084223
|
epc1b
|
enhancer of polycomb homolog 1 (Drosophila) b |
| chr1_-_40994259 | 14.65 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr14_-_25577094 | 14.64 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr4_+_7888047 | 14.62 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr3_+_15907297 | 14.61 |
ENSDART00000139206
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr7_+_23907692 | 14.57 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr20_+_50956369 | 14.57 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr13_+_35745572 | 14.56 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr21_+_30563115 | 14.55 |
ENSDART00000028566
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr6_+_12853655 | 14.53 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr9_-_35557397 | 14.48 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr16_+_43152727 | 14.33 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr1_+_36436936 | 14.28 |
ENSDART00000124112
|
pou4f2
|
POU class 4 homeobox 2 |
| chr17_+_23937262 | 14.27 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr3_-_36944749 | 14.27 |
ENSDART00000154501
|
cntnap1
|
contactin associated protein 1 |
| chr22_-_29922872 | 14.24 |
ENSDART00000020249
|
dusp5
|
dual specificity phosphatase 5 |
| chr21_+_41697552 | 14.17 |
ENSDART00000169511
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr19_+_26718074 | 14.11 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr3_+_26044969 | 14.09 |
ENSDART00000133523
|
hmgxb4a
|
HMG box domain containing 4a |
| chr14_+_25465346 | 14.03 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr22_-_25033105 | 13.97 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr13_+_36146415 | 13.95 |
ENSDART00000140301
|
TTC9
|
si:ch211-259k16.3 |
| chr23_-_8373676 | 13.92 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr16_-_13662514 | 13.88 |
ENSDART00000146348
|
shisa7a
|
shisa family member 7a |
| chr3_+_29714775 | 13.83 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr17_-_20979077 | 13.80 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr9_-_54840124 | 13.74 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr17_+_41463942 | 13.68 |
ENSDART00000075331
|
insm1b
|
insulinoma-associated 1b |
| chr5_-_24869213 | 13.58 |
ENSDART00000112287
ENSDART00000144635 |
gas2l1
|
growth arrest-specific 2 like 1 |
| chr5_-_67878064 | 13.46 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr10_+_32561317 | 13.42 |
ENSDART00000109029
|
map6a
|
microtubule-associated protein 6a |
| chr10_+_23022263 | 13.32 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr10_+_17850934 | 13.31 |
ENSDART00000113666
ENSDART00000145936 |
phf24
|
PHD finger protein 24 |
| chr13_-_32906265 | 13.30 |
ENSDART00000113823
ENSDART00000171114 ENSDART00000180035 ENSDART00000137570 |
si:dkey-18j18.3
|
si:dkey-18j18.3 |
| chr24_-_6546479 | 13.28 |
ENSDART00000160538
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr19_+_342094 | 13.25 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr8_+_28066063 | 13.24 |
ENSDART00000078533
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr6_-_15604157 | 13.23 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr8_+_23916647 | 13.12 |
ENSDART00000143152
|
cpne5a
|
copine Va |
| chr23_-_3703569 | 13.07 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr10_-_34772211 | 13.06 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr6_+_39222598 | 13.03 |
ENSDART00000154991
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr19_-_31007417 | 13.02 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr7_-_52963493 | 12.95 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr8_-_34052019 | 12.95 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr4_-_12007404 | 12.71 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr23_+_44741500 | 12.68 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr21_-_25522906 | 12.62 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr1_-_22834824 | 12.55 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr4_+_9279784 | 12.51 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr7_-_28147838 | 12.41 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr4_+_4849789 | 12.33 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr14_+_11103718 | 12.29 |
ENSDART00000161311
|
nexmifb
|
neurite extension and migration factor b |
| chr23_-_30787932 | 12.11 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr23_-_12014931 | 12.07 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr1_+_32521469 | 12.06 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr1_+_24076243 | 12.04 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr4_+_10366532 | 12.01 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr13_-_29424454 | 11.95 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr7_-_69983462 | 11.92 |
ENSDART00000123380
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr20_+_88168 | 11.90 |
ENSDART00000149283
|
zgc:112001
|
zgc:112001 |
| chr3_+_35005730 | 11.85 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr1_-_53756851 | 11.84 |
ENSDART00000122445
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr16_-_24518027 | 11.77 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr10_+_21807497 | 11.71 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr14_+_45259644 | 11.48 |
ENSDART00000168270
|
CABZ01048053.1
|
|
| chr3_-_6767440 | 11.42 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr3_+_15907458 | 11.41 |
ENSDART00000163525
|
mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr23_-_29751730 | 11.39 |
ENSDART00000056865
|
ctnnbip1
|
catenin, beta interacting protein 1 |
| chr7_-_39203799 | 11.33 |
ENSDART00000173727
|
chrm4a
|
cholinergic receptor, muscarinic 4a |
| chr20_-_40451115 | 11.29 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr3_+_19914332 | 11.27 |
ENSDART00000078982
|
vat1
|
vesicle amine transport 1 |
| chr5_-_10946232 | 11.27 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr7_+_14632157 | 11.26 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr9_+_44722205 | 11.24 |
ENSDART00000086176
ENSDART00000145271 ENSDART00000132696 |
nckap1
|
NCK-associated protein 1 |
| chr13_-_27767330 | 11.23 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr15_+_28482862 | 11.18 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr8_+_26329482 | 11.14 |
ENSDART00000048676
|
prkar2aa
|
protein kinase, cAMP-dependent, regulatory, type II, alpha A |
| chr4_-_22671469 | 11.14 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr25_-_21092222 | 11.11 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr10_+_22381802 | 11.05 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr1_-_55873178 | 11.04 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr12_+_12112384 | 10.98 |
ENSDART00000152431
|
grid1b
|
glutamate receptor, ionotropic, delta 1b |
| chr8_+_39760258 | 10.95 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr17_+_52822831 | 10.80 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr21_+_11468642 | 10.77 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr6_+_38427357 | 10.75 |
ENSDART00000148678
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr3_+_37824268 | 10.74 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr1_+_9004719 | 10.60 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr2_+_23222939 | 10.57 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr4_+_10888762 | 10.55 |
ENSDART00000136049
|
syt10
|
synaptotagmin X |
| chr1_-_10647307 | 10.51 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr23_-_36305874 | 10.47 |
ENSDART00000147598
ENSDART00000146986 ENSDART00000086985 ENSDART00000133259 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr4_-_24019711 | 10.44 |
ENSDART00000077926
|
celf2
|
cugbp, Elav-like family member 2 |
| chr7_+_41748693 | 10.41 |
ENSDART00000174379
ENSDART00000052168 |
hrh3
|
histamine receptor H3 |
| chr11_+_45153104 | 10.40 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr9_-_1703761 | 10.38 |
ENSDART00000144822
ENSDART00000137210 ENSDART00000135273 |
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr16_+_41015163 | 10.28 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr20_-_23226453 | 10.28 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr14_-_40389699 | 10.25 |
ENSDART00000181581
ENSDART00000173398 |
pcdh19
|
protocadherin 19 |
| chr3_+_46628885 | 10.22 |
ENSDART00000006602
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
| chr3_-_50177658 | 10.16 |
ENSDART00000135309
|
zgc:114118
|
zgc:114118 |
| chr11_-_45434959 | 10.09 |
ENSDART00000173106
ENSDART00000172767 ENSDART00000172933 ENSDART00000172986 |
rfc4
|
replication factor C (activator 1) 4 |
| chr2_+_23701613 | 10.02 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr6_-_30839763 | 9.97 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr1_-_54997746 | 9.97 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr13_-_51973562 | 9.94 |
ENSDART00000166701
|
slc30a10
|
solute carrier family 30, member 10 |
| chr6_+_36844127 | 9.92 |
ENSDART00000020505
|
chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr3_+_54168007 | 9.86 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr14_-_2352384 | 9.82 |
ENSDART00000170666
|
si:ch73-233f7.7
|
si:ch73-233f7.7 |
| chr3_+_24986145 | 9.81 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr2_+_48303142 | 9.81 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr17_+_24109012 | 9.78 |
ENSDART00000156251
|
ehbp1
|
EH domain binding protein 1 |
| chr16_+_68069 | 9.77 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr4_-_25064510 | 9.65 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr6_+_27090800 | 9.65 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr12_+_42436920 | 9.64 |
ENSDART00000177303
|
ebf3a
|
early B cell factor 3a |
| chr15_-_31516558 | 9.60 |
ENSDART00000156427
ENSDART00000156072 ENSDART00000156047 |
hmgb1b
|
high mobility group box 1b |
| chr8_+_6967108 | 9.51 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr10_-_28835771 | 9.49 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
| chr25_-_3503164 | 9.43 |
ENSDART00000191477
ENSDART00000186345 ENSDART00000180199 |
PRKAR2B
|
si:ch211-272n13.7 |
| chr20_-_20610812 | 9.41 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr17_-_11329959 | 9.39 |
ENSDART00000015418
|
irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr16_+_22618620 | 9.35 |
ENSDART00000185728
ENSDART00000041625 |
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr17_-_23674495 | 9.22 |
ENSDART00000122209
|
ptena
|
phosphatase and tensin homolog A |
| chr11_-_15001639 | 9.21 |
ENSDART00000170627
ENSDART00000168222 ENSDART00000171210 ENSDART00000163518 ENSDART00000168180 ENSDART00000157970 ENSDART00000169113 ENSDART00000180049 ENSDART00000188961 ENSDART00000183952 ENSDART00000193904 ENSDART00000193450 ENSDART00000186508 ENSDART00000169476 ENSDART00000162938 |
elavl1b
|
ELAV like RNA binding protein 1b |
| chr14_-_27297123 | 9.21 |
ENSDART00000173423
|
pcdh11
|
protocadherin 11 |
| chr6_-_26559921 | 9.17 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr2_+_5621529 | 9.16 |
ENSDART00000144187
|
fgf12a
|
fibroblast growth factor 12a |
| chr17_+_19630272 | 9.13 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr6_+_4539953 | 9.12 |
ENSDART00000025031
|
pou4f1
|
POU class 4 homeobox 1 |
| chr12_-_23009312 | 9.10 |
ENSDART00000111801
|
mkxa
|
mohawk homeobox a |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfdp1a+tfdp1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 26.2 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 7.1 | 28.3 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 6.0 | 18.0 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 4.9 | 14.8 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 4.9 | 14.6 | GO:0072579 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 4.7 | 14.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 4.6 | 23.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 4.1 | 20.7 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 4.0 | 12.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 4.0 | 23.8 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 3.9 | 19.3 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 3.7 | 14.8 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 3.5 | 17.7 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 3.5 | 10.4 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 3.4 | 17.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 3.4 | 10.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 3.4 | 30.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 3.3 | 29.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 3.1 | 9.4 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to metal ion(GO:0097501) stress response to zinc ion(GO:1990359) |
| 3.0 | 45.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 2.9 | 26.0 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 2.7 | 46.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 2.6 | 7.9 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 2.5 | 17.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 2.5 | 7.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 2.4 | 24.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 2.3 | 9.2 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 2.3 | 6.8 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 2.3 | 13.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 2.2 | 70.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 2.2 | 6.5 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 2.0 | 11.8 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 1.9 | 1.9 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 1.9 | 7.6 | GO:0015677 | copper ion import(GO:0015677) |
| 1.9 | 11.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.9 | 16.7 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 1.8 | 7.1 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 1.8 | 8.9 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 1.8 | 5.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 1.7 | 5.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.7 | 6.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.7 | 28.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 1.7 | 14.9 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 1.6 | 12.9 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 1.6 | 4.8 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 1.6 | 6.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 1.6 | 4.7 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 1.6 | 6.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.5 | 19.9 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 1.5 | 7.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 1.5 | 4.5 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 1.4 | 4.3 | GO:0043586 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 1.4 | 4.3 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 1.4 | 14.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 1.4 | 14.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 1.4 | 7.1 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 1.4 | 9.6 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 1.4 | 4.1 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 1.4 | 9.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 1.3 | 13.9 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 1.2 | 13.7 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 1.2 | 11.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 1.2 | 12.0 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 1.1 | 5.7 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 1.1 | 7.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.1 | 7.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 1.1 | 62.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 1.1 | 22.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 1.0 | 11.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 1.0 | 16.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 1.0 | 53.2 | GO:0051588 | regulation of neurotransmitter transport(GO:0051588) |
| 1.0 | 14.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.0 | 11.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 1.0 | 11.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 1.0 | 13.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.9 | 33.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.9 | 4.7 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.9 | 4.6 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.9 | 5.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.9 | 22.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.9 | 2.6 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.9 | 3.5 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.8 | 5.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.8 | 13.5 | GO:0036065 | fucosylation(GO:0036065) |
| 0.8 | 39.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.8 | 3.3 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.8 | 9.0 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.8 | 2.5 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.8 | 4.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.8 | 8.5 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.8 | 1.5 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.8 | 6.2 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.8 | 2.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.8 | 7.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.8 | 5.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.7 | 4.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.7 | 3.0 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.7 | 10.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.7 | 5.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.7 | 21.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.7 | 7.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.7 | 2.2 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.7 | 11.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.7 | 3.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.7 | 2.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.7 | 11.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.7 | 7.7 | GO:0060419 | cardiac muscle tissue growth(GO:0055017) cardiac muscle cell proliferation(GO:0060038) heart growth(GO:0060419) |
| 0.7 | 5.5 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.7 | 2.0 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.7 | 12.7 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.7 | 16.0 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.7 | 9.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.7 | 3.3 | GO:0055016 | hypochord development(GO:0055016) |
| 0.6 | 3.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.6 | 6.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.6 | 8.2 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.6 | 1.9 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.6 | 24.2 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.6 | 7.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.6 | 7.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.6 | 3.6 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.6 | 1.8 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.6 | 3.0 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.6 | 13.1 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.6 | 11.3 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.6 | 3.5 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.6 | 6.4 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.6 | 2.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.6 | 4.5 | GO:0071632 | thalamus development(GO:0021794) optomotor response(GO:0071632) |
| 0.6 | 13.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.6 | 5.5 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.6 | 2.2 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.5 | 1.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.5 | 1.6 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.5 | 22.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.5 | 8.9 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.5 | 8.4 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.5 | 5.2 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.5 | 3.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.5 | 10.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.5 | 13.3 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.5 | 7.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.5 | 4.3 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.5 | 6.7 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.5 | 2.4 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.5 | 1.4 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.5 | 2.3 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.5 | 9.1 | GO:0001505 | regulation of neurotransmitter levels(GO:0001505) |
| 0.5 | 2.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.5 | 9.5 | GO:0048883 | neuromast primordium migration(GO:0048883) posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.4 | 7.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.4 | 107.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 15.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.4 | 5.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 2.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.4 | 5.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 5.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.4 | 8.6 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.4 | 3.9 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.4 | 6.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.4 | 5.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 2.9 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 4.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.4 | 12.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.4 | 4.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.4 | 6.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.4 | 23.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.4 | 4.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.4 | 6.6 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.4 | 3.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.4 | 11.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.4 | 2.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.4 | 15.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 2.8 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.3 | 1.7 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.3 | 1.7 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.3 | 19.0 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.3 | 25.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.3 | 3.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 5.6 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.3 | 0.7 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.3 | 4.8 | GO:0072176 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
| 0.3 | 6.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.3 | 2.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 4.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.3 | 3.1 | GO:0021694 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 1.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 11.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.3 | 5.9 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.3 | 1.1 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.3 | 5.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.3 | 45.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.3 | 1.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 4.1 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.2 | 4.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.2 | 9.1 | GO:0035336 | long-chain fatty acid metabolic process(GO:0001676) long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.2 | 4.4 | GO:0099587 | inorganic cation import into cell(GO:0098659) inorganic ion import into cell(GO:0099587) |
| 0.2 | 1.2 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.2 | 1.6 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 20.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.2 | 19.2 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.2 | 2.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 10.2 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.2 | 5.8 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.2 | 4.9 | GO:0021537 | telencephalon development(GO:0021537) |
| 0.2 | 3.4 | GO:0006003 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 4.6 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.2 | 2.5 | GO:0050890 | cognition(GO:0050890) |
| 0.2 | 4.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 12.8 | GO:0042552 | myelination(GO:0042552) |
| 0.2 | 4.0 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.2 | 10.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 3.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.2 | 25.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.2 | 5.9 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.2 | 22.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.2 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 15.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.2 | 0.7 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.2 | 1.1 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.2 | 1.0 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 8.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.2 | 1.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.2 | 2.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 3.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 1.6 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.2 | 0.8 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.2 | 1.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 3.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.8 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.6 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 16.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 4.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 5.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 11.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 5.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.1 | 2.3 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 1.7 | GO:0016125 | sterol metabolic process(GO:0016125) |
| 0.1 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 4.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.7 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 1.0 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.7 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) |
| 0.1 | 2.8 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 0.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 1.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 2.8 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 1.3 | GO:0036353 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.1 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.1 | 3.6 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 1.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 2.6 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 0.4 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 20.3 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.1 | 15.2 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 3.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 8.4 | GO:0007051 | spindle organization(GO:0007051) |
| 0.1 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 4.7 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 10.5 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 10.2 | GO:0048675 | axon extension(GO:0048675) |
| 0.1 | 1.5 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 4.4 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 3.8 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.1 | 2.2 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 11.5 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 1.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 4.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 23.9 | GO:0007420 | brain development(GO:0007420) |
| 0.1 | 3.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 1.7 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.1 | 2.1 | GO:0034111 | negative regulation of homotypic cell-cell adhesion(GO:0034111) negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.1 | 0.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 2.9 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.3 | GO:0051899 | membrane depolarization(GO:0051899) membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 4.5 | GO:0046578 | regulation of Ras protein signal transduction(GO:0046578) |
| 0.0 | 3.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 1.8 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 3.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.1 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 1.3 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.7 | GO:0007033 | vacuole organization(GO:0007033) |
| 0.0 | 8.5 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.7 | 26.2 | GO:0072380 | TRC complex(GO:0072380) |
| 7.3 | 22.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 5.8 | 23.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 3.7 | 11.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 3.2 | 142.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 2.7 | 5.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 2.4 | 19.0 | GO:0033010 | paranodal junction(GO:0033010) |
| 2.2 | 8.9 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 2.2 | 53.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 1.9 | 11.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.8 | 20.1 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 1.6 | 11.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 1.5 | 6.0 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.4 | 8.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.4 | 9.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 1.4 | 4.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 1.3 | 8.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 1.3 | 12.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 1.2 | 11.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.2 | 16.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 1.0 | 4.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 1.0 | 7.9 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 1.0 | 31.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.9 | 5.6 | GO:0000938 | GARP complex(GO:0000938) |
| 0.9 | 8.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.9 | 3.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.8 | 19.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.8 | 14.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.8 | 3.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.8 | 8.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.8 | 25.8 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.7 | 4.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.7 | 7.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.7 | 4.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.7 | 16.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.6 | 25.1 | GO:0016605 | PML body(GO:0016605) |
| 0.6 | 9.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.6 | 15.6 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.6 | 117.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.6 | 10.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.5 | 20.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.5 | 20.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.5 | 28.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.5 | 7.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.4 | 1.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.4 | 5.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.4 | 1.7 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 4.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.4 | 9.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 5.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 3.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.4 | 11.2 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.4 | 11.1 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.4 | 11.9 | GO:0060293 | germ plasm(GO:0060293) |
| 0.4 | 13.1 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.4 | 4.3 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 8.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 1.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.3 | 15.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.3 | 5.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 6.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 20.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 5.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 1.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 15.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 15.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 1.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 11.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 5.1 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.2 | 3.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 12.3 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.2 | 48.9 | GO:0030424 | axon(GO:0030424) |
| 0.2 | 5.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 9.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 0.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 6.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.2 | 12.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 72.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.2 | 1.6 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 5.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 10.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 7.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 7.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 2.9 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 7.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 132.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 3.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 2.9 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.1 | 1.6 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 9.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 18.3 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 42.3 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 3.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 3.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.1 | 22.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 11.3 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 13.2 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 6.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 2.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 16.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 2.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.5 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.2 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 3.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.7 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 64.7 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 3.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 74.1 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 42.5 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 23.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 5.5 | 32.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 5.2 | 26.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 5.0 | 75.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 5.0 | 29.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 4.9 | 14.6 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 4.3 | 17.2 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 4.1 | 16.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 3.2 | 16.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 3.0 | 14.8 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 2.9 | 40.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 2.8 | 22.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.4 | 17.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 2.3 | 6.8 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 2.1 | 8.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 2.1 | 6.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 2.1 | 6.4 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 1.9 | 13.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.9 | 7.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.9 | 9.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 1.9 | 5.7 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 1.9 | 11.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 1.9 | 11.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 1.8 | 5.4 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 1.7 | 5.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 1.7 | 6.7 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 1.6 | 11.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.5 | 13.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 1.5 | 20.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 1.5 | 10.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 1.5 | 18.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.5 | 59.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.5 | 5.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 1.4 | 8.7 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.4 | 4.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.4 | 9.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 1.4 | 4.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.2 | 8.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.1 | 8.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.1 | 5.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 1.1 | 16.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.1 | 7.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 1.0 | 9.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 1.0 | 11.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 1.0 | 14.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.0 | 6.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.0 | 7.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.0 | 5.9 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.0 | 46.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 1.0 | 14.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.9 | 47.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.9 | 12.0 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.9 | 5.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.9 | 2.7 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.9 | 13.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.8 | 6.6 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.8 | 3.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 13.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.8 | 7.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.8 | 66.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.8 | 8.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.8 | 7.0 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.8 | 25.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.8 | 6.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.7 | 32.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.7 | 10.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.7 | 4.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.7 | 2.2 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.7 | 13.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.7 | 10.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.7 | 11.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.7 | 33.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.7 | 4.8 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.7 | 5.5 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.7 | 4.1 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.7 | 23.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.7 | 6.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.7 | 8.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.7 | 15.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.6 | 7.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.6 | 26.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.6 | 5.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 12.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.6 | 23.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.6 | 7.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.6 | 4.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.6 | 6.4 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.6 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.6 | 13.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.6 | 9.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.6 | 19.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.5 | 3.2 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.5 | 5.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.5 | 2.6 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.5 | 8.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 10.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 4.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.5 | 11.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.5 | 5.5 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.5 | 3.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.5 | 18.8 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.5 | 4.3 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.5 | 9.5 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.5 | 31.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.5 | 6.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.5 | 7.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.5 | 1.8 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.5 | 3.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.4 | 15.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 7.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.4 | 3.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.4 | 1.3 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.4 | 24.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.4 | 7.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 8.9 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.4 | 2.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.4 | 1.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.4 | 3.5 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.4 | 23.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.4 | 7.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 7.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.4 | 3.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 2.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 15.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.3 | 4.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 1.4 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.3 | 2.4 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 5.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.3 | 4.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 1.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 2.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 5.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 16.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 7.2 | GO:0015464 | acetylcholine receptor activity(GO:0015464) acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 1.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.3 | 3.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.3 | 0.9 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 6.9 | GO:0009931 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 6.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.3 | 5.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.3 | 4.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 4.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.3 | 16.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 2.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 3.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 7.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.2 | 2.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 6.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 25.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.2 | 9.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.2 | 1.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 14.8 | GO:0051287 | NAD binding(GO:0051287) |
| 0.2 | 1.0 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.2 | 0.8 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.2 | 6.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 2.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 5.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 182.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 4.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.7 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.2 | 1.8 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 7.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 2.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 2.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 22.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.2 | 3.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 15.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 0.7 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.2 | 22.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.2 | 5.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 6.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 50.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 5.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 3.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 3.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 20.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 7.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 40.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.7 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 6.1 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 4.6 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 3.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 3.8 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 4.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 3.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 3.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 18.2 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.1 | 2.9 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 3.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 13.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 3.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 5.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 2.2 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 1.0 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 24.6 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.8 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 4.7 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 52.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 2.4 | 50.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 1.3 | 26.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.7 | 5.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.6 | 5.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 5.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 2.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.4 | 10.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.4 | 9.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 4.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.4 | 1.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 27.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.4 | 18.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 7.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.4 | 2.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.3 | 3.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.3 | 11.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 5.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 7.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 23.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 10.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.2 | 4.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 1.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 1.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 3.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 4.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 1.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 11.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 4.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 4.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 5.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 2.1 | 25.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 1.8 | 34.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.8 | 28.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.7 | 25.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.7 | 15.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 1.7 | 6.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.7 | 18.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.6 | 22.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 1.4 | 17.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 1.3 | 11.9 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 1.2 | 12.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 1.2 | 5.8 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 1.0 | 5.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 1.0 | 7.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.8 | 7.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.8 | 10.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.8 | 4.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.7 | 7.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.7 | 11.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.6 | 18.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.6 | 16.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.6 | 4.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.6 | 7.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.5 | 6.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.5 | 7.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.5 | 10.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.4 | 7.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 2.1 | REACTOME LAGGING STRAND SYNTHESIS | Genes involved in Lagging Strand Synthesis |
| 0.4 | 13.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 9.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.4 | 12.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.3 | 1.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.3 | 2.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 4.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 1.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.3 | 9.2 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.3 | 19.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.3 | 3.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 16.6 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.3 | 4.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 18.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 6.4 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 5.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 17.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 20.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 4.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 3.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 6.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 2.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 0.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 4.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 5.6 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |