Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
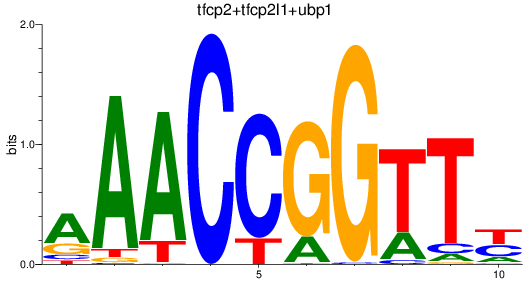
Results for tfcp2+tfcp2l1+ubp1
Z-value: 2.16
Transcription factors associated with tfcp2+tfcp2l1+ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ubp1
|
ENSDARG00000018000 | upstream binding protein 1 |
|
tfcp2l1
|
ENSDARG00000029497 | transcription factor CP2-like 1 |
|
tfcp2
|
ENSDARG00000060306 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfcp2 | dr11_v1_chr23_-_33680265_33680265 | 0.38 | 1.9e-04 | Click! |
| tfcp2l1 | dr11_v1_chr9_+_38292947_38292947 | -0.36 | 3.3e-04 | Click! |
| ubp1 | dr11_v1_chr19_+_42806812_42806814 | 0.30 | 3.3e-03 | Click! |
Activity profile of tfcp2+tfcp2l1+ubp1 motif
Sorted Z-values of tfcp2+tfcp2l1+ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_27647845 | 36.85 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr10_-_15048781 | 36.37 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr5_+_39667272 | 23.22 |
ENSDART00000085388
|
bmp3
|
bone morphogenetic protein 3 |
| chr21_+_25688388 | 22.67 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr12_-_28794957 | 21.72 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr5_+_15203421 | 21.53 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr16_+_23961276 | 19.43 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr10_-_36793412 | 18.96 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr24_+_20960216 | 18.95 |
ENSDART00000133008
|
si:ch211-161h7.8
|
si:ch211-161h7.8 |
| chr8_+_36142734 | 17.87 |
ENSDART00000159361
ENSDART00000161194 |
mhc2b
|
major histocompatibility complex class II integral membrane beta chain gene |
| chr9_-_45602978 | 17.62 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr24_+_38671054 | 17.21 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr22_-_6404807 | 17.04 |
ENSDART00000149399
ENSDART00000063420 |
si:rp71-1i20.2
|
si:rp71-1i20.2 |
| chr8_-_3312384 | 16.55 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr7_+_20503344 | 16.38 |
ENSDART00000157699
|
si:dkey-19b23.12
|
si:dkey-19b23.12 |
| chr5_+_26204561 | 15.99 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr10_+_36662640 | 15.75 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr20_+_26916639 | 15.54 |
ENSDART00000077787
|
serpinb1l2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 2 |
| chr17_-_2039511 | 15.24 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr12_-_46145635 | 14.93 |
ENSDART00000074682
|
zgc:153932
|
zgc:153932 |
| chr25_+_10416583 | 14.92 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr22_+_22021936 | 14.77 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr9_-_443451 | 14.65 |
ENSDART00000165642
|
si:dkey-11f4.14
|
si:dkey-11f4.14 |
| chr23_+_18779043 | 14.47 |
ENSDART00000158267
|
emp3b
|
epithelial membrane protein 3b |
| chr5_-_37116265 | 14.33 |
ENSDART00000057613
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr15_+_22014029 | 13.97 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr2_-_15563194 | 12.93 |
ENSDART00000049589
|
col11a1b
|
collagen, type XI, alpha 1b |
| chr12_-_46252062 | 12.84 |
ENSDART00000153223
|
si:ch211-226h7.5
|
si:ch211-226h7.5 |
| chr15_+_46329149 | 12.72 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr17_-_37395460 | 12.35 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr4_+_76671012 | 12.34 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr15_-_34892664 | 12.17 |
ENSDART00000153787
ENSDART00000099721 |
rnf183
|
ring finger protein 183 |
| chr16_-_42004544 | 11.69 |
ENSDART00000034544
|
caspa
|
caspase a |
| chr5_+_30741730 | 11.67 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr13_-_25774183 | 11.49 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr7_+_73670137 | 11.47 |
ENSDART00000050357
|
btr12
|
bloodthirsty-related gene family, member 12 |
| chr8_+_52479026 | 11.46 |
ENSDART00000162885
|
FO704661.1
|
Danio rerio gamma-glutamyl hydrolase (LOC553228), mRNA. |
| chr22_+_22004082 | 10.94 |
ENSDART00000148375
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr3_-_26204867 | 10.93 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr4_-_77979432 | 10.89 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr14_-_25956804 | 10.66 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr12_-_4540564 | 10.66 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr21_-_32436679 | 10.65 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr17_+_51743908 | 10.54 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr6_-_35401282 | 10.46 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr14_+_33329420 | 10.26 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr12_+_46634736 | 10.25 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr3_-_32590164 | 10.25 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr16_-_6424816 | 10.22 |
ENSDART00000164864
ENSDART00000141860 |
mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr22_-_10502780 | 10.16 |
ENSDART00000136961
|
ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr25_+_30196039 | 10.05 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr3_-_53092509 | 10.02 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr16_+_51180938 | 10.02 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr7_+_19835569 | 10.01 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr5_+_58665648 | 9.91 |
ENSDART00000167481
|
zgc:194948
|
zgc:194948 |
| chr20_+_16881883 | 9.71 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr12_-_16990896 | 9.27 |
ENSDART00000152402
|
ifit12
|
interferon-induced protein with tetratricopeptide repeats 12 |
| chr9_-_3400727 | 9.17 |
ENSDART00000183979
ENSDART00000111386 |
dlx2a
|
distal-less homeobox 2a |
| chr6_-_24103666 | 9.12 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr12_+_38929663 | 8.94 |
ENSDART00000156334
|
si:dkey-239b22.1
|
si:dkey-239b22.1 |
| chr15_+_12429206 | 8.84 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr12_+_38878830 | 8.67 |
ENSDART00000156926
|
si:ch211-39f2.3
|
si:ch211-39f2.3 |
| chr7_+_57088920 | 8.54 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr13_+_669177 | 8.50 |
ENSDART00000190085
|
CU462915.1
|
|
| chr21_+_19648814 | 8.40 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr5_-_23855447 | 8.25 |
ENSDART00000051541
|
gbgt1l3
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 3 |
| chr15_+_41919484 | 8.12 |
ENSDART00000099821
ENSDART00000146246 |
nlrp16
|
NACHT, LRR and PYD domains-containing protein 16 |
| chr21_-_13668358 | 8.11 |
ENSDART00000180323
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr21_-_42831033 | 7.96 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr22_+_9806867 | 7.95 |
ENSDART00000137182
ENSDART00000106103 |
si:ch211-236g6.1
|
si:ch211-236g6.1 |
| chr21_+_25625026 | 7.92 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr3_+_6469754 | 7.88 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr22_-_3299355 | 7.80 |
ENSDART00000190993
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr9_-_11550711 | 7.55 |
ENSDART00000093343
|
fev
|
FEV (ETS oncogene family) |
| chr6_+_23810529 | 7.54 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr8_-_46457233 | 7.54 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr18_+_27571448 | 7.53 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr13_-_40411908 | 7.53 |
ENSDART00000057094
ENSDART00000150091 |
nkx2.3
|
NK2 homeobox 3 |
| chr5_+_36599107 | 7.28 |
ENSDART00000097677
ENSDART00000138147 |
micu2
|
mitochondrial calcium uptake 2 |
| chr18_-_40773413 | 7.26 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr22_+_19552987 | 7.21 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr5_+_20319519 | 7.04 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr16_+_29509133 | 6.99 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr14_+_33329761 | 6.72 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr2_-_47620806 | 6.70 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr1_-_21321482 | 6.63 |
ENSDART00000054440
|
tmem144a
|
transmembrane protein 144a |
| chr8_-_28349859 | 6.58 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr5_+_21891305 | 6.52 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr22_-_15587360 | 6.45 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr15_+_13984879 | 6.44 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr1_-_56556326 | 6.25 |
ENSDART00000188760
|
CABZ01059408.1
|
|
| chr2_-_43151358 | 6.20 |
ENSDART00000061170
ENSDART00000056161 |
ftr15
|
finTRIM family, member 15 |
| chr16_+_29586004 | 6.08 |
ENSDART00000149520
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr16_+_29586468 | 6.08 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr20_-_39367895 | 5.97 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr23_-_26252103 | 5.92 |
ENSDART00000160873
|
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr3_+_5083407 | 5.68 |
ENSDART00000146883
|
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr8_-_32805214 | 5.63 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr7_+_57089354 | 5.63 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr13_+_24671481 | 5.42 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr10_+_39084354 | 5.40 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr13_+_18533005 | 5.32 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr14_+_7048930 | 5.28 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr10_+_44692272 | 5.25 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr23_+_7471072 | 5.22 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr22_-_4778018 | 5.19 |
ENSDART00000143968
|
si:ch73-256j6.5
|
si:ch73-256j6.5 |
| chr10_+_36458563 | 5.11 |
ENSDART00000077008
|
alox5ap
|
arachidonate 5-lipoxygenase-activating protein |
| chr22_-_3299100 | 5.03 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr12_+_30234209 | 5.02 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr7_+_57725708 | 5.01 |
ENSDART00000056466
ENSDART00000142259 ENSDART00000166198 |
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr4_+_78058635 | 4.94 |
ENSDART00000162631
ENSDART00000191619 |
CABZ01075274.1
|
|
| chr4_-_77636185 | 4.89 |
ENSDART00000152915
ENSDART00000131964 |
si:dkey-61p9.9
|
si:dkey-61p9.9 |
| chr7_-_52417777 | 4.85 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr5_+_50953240 | 4.80 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr22_-_17631675 | 4.74 |
ENSDART00000132565
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr15_-_28904371 | 4.74 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr6_+_58915889 | 4.73 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr2_+_44571200 | 4.72 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr6_+_13506841 | 4.70 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr8_-_13029297 | 4.65 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr3_-_34528306 | 4.64 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr14_-_31087830 | 4.59 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr13_-_36798204 | 4.58 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr25_+_10923100 | 4.49 |
ENSDART00000157055
|
si:ch211-147g22.7
|
si:ch211-147g22.7 |
| chr16_+_5597600 | 4.46 |
ENSDART00000017307
|
zgc:91890
|
zgc:91890 |
| chr13_+_31321297 | 4.45 |
ENSDART00000143308
|
antxr1d
|
anthrax toxin receptor 1d |
| chr3_-_29977495 | 4.41 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr23_-_3511630 | 4.38 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr12_+_30233977 | 4.31 |
ENSDART00000188135
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr8_+_52377516 | 4.26 |
ENSDART00000115398
|
arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr17_-_43677471 | 4.23 |
ENSDART00000137015
|
egr2a
|
early growth response 2a |
| chr5_+_72152813 | 4.18 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr4_-_26035770 | 4.11 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr7_-_24193051 | 4.10 |
ENSDART00000169737
ENSDART00000171085 |
si:ch211-216p19.6
|
si:ch211-216p19.6 |
| chr12_+_22657925 | 4.10 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr17_+_15213496 | 4.08 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr7_-_56606752 | 4.08 |
ENSDART00000138714
|
sult5a1
|
sulfotransferase family 5A, member 1 |
| chr5_+_43807003 | 4.06 |
ENSDART00000097625
|
zgc:158640
|
zgc:158640 |
| chr19_-_2861444 | 4.00 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr6_+_41452979 | 3.99 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr21_-_14310159 | 3.98 |
ENSDART00000155097
|
si:ch211-196i2.1
|
si:ch211-196i2.1 |
| chr6_-_48094342 | 3.90 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr3_-_36846496 | 3.88 |
ENSDART00000055237
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr4_+_6032640 | 3.85 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr1_-_46401385 | 3.80 |
ENSDART00000150029
|
atp11a
|
ATPase phospholipid transporting 11A |
| chr5_-_67750907 | 3.80 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr20_-_16852 | 3.79 |
ENSDART00000129277
ENSDART00000148717 |
zgc:174972
|
zgc:174972 |
| chr10_-_1523253 | 3.79 |
ENSDART00000179510
ENSDART00000176548 ENSDART00000180368 ENSDART00000185270 |
WDR70
|
WD repeat domain 70 |
| chr15_-_34865952 | 3.72 |
ENSDART00000186868
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr21_-_30166097 | 3.71 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr7_-_54430505 | 3.69 |
ENSDART00000167905
|
ano1
|
anoctamin 1, calcium activated chloride channel |
| chr14_+_17376940 | 3.68 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr14_-_31489410 | 3.64 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr19_+_31576849 | 3.64 |
ENSDART00000134107
ENSDART00000088401 |
acot13
|
acyl-CoA thioesterase 13 |
| chr8_+_25342896 | 3.57 |
ENSDART00000129032
|
CR847543.1
|
|
| chr18_+_47313715 | 3.53 |
ENSDART00000138806
|
barx2
|
BARX homeobox 2 |
| chr7_+_19483277 | 3.52 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr7_+_66739166 | 3.45 |
ENSDART00000154634
|
mrvi1
|
murine retrovirus integration site 1 homolog |
| chr21_-_8085635 | 3.44 |
ENSDART00000082790
|
si:dkey-163m14.2
|
si:dkey-163m14.2 |
| chr4_-_20222182 | 3.44 |
ENSDART00000132464
|
gsap
|
gamma-secretase activating protein |
| chr15_-_5901514 | 3.43 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr22_-_16275236 | 3.40 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr8_+_27555314 | 3.38 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr19_+_23308419 | 3.35 |
ENSDART00000141558
|
irgf2
|
immunity-related GTPase family, f2 |
| chr15_-_452347 | 3.34 |
ENSDART00000115233
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr10_+_29204581 | 3.31 |
ENSDART00000148503
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr2_-_42035250 | 3.29 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr12_-_34035364 | 3.29 |
ENSDART00000087065
|
timp2a
|
TIMP metallopeptidase inhibitor 2a |
| chr11_+_25112269 | 3.27 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr6_-_49526510 | 3.26 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr20_-_25631256 | 3.25 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr8_-_29658171 | 3.25 |
ENSDART00000147755
|
mpeg1.3
|
macrophage expressed 1, tandem duplicate 3 |
| chr8_-_52715911 | 3.21 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr11_+_25111846 | 3.18 |
ENSDART00000128705
ENSDART00000190058 |
ndrg3a
|
ndrg family member 3a |
| chr21_+_15375785 | 3.18 |
ENSDART00000147666
|
si:dkey-11o15.8
|
si:dkey-11o15.8 |
| chr12_+_48841182 | 3.14 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr2_-_7711522 | 3.13 |
ENSDART00000163175
|
dcun1d1
|
DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) |
| chr8_-_11067079 | 3.12 |
ENSDART00000181986
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr8_-_418997 | 3.09 |
ENSDART00000166683
|
pggt1b
|
protein geranylgeranyltransferase type I, beta subunit |
| chr4_+_18963822 | 3.05 |
ENSDART00000066975
ENSDART00000066973 |
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr13_-_11679486 | 3.03 |
ENSDART00000108694
|
si:ch211-195e19.1
|
si:ch211-195e19.1 |
| chr24_+_5935377 | 2.98 |
ENSDART00000191989
ENSDART00000185932 ENSDART00000131768 |
abi1a
|
abl-interactor 1a |
| chr18_+_17725410 | 2.97 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr20_+_53181017 | 2.96 |
ENSDART00000189692
ENSDART00000177109 |
FIG4
|
FIG4 phosphoinositide 5-phosphatase |
| chr14_+_20941534 | 2.95 |
ENSDART00000185616
|
zgc:66433
|
zgc:66433 |
| chr3_-_26806032 | 2.87 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr5_-_38438613 | 2.86 |
ENSDART00000131912
|
pld2
|
phospholipase D2 |
| chr12_-_10476448 | 2.82 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr22_+_19640309 | 2.79 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr15_-_34668485 | 2.78 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr23_-_270847 | 2.76 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr14_+_45350008 | 2.75 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr7_-_34256374 | 2.73 |
ENSDART00000075176
|
dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr13_+_18471546 | 2.70 |
ENSDART00000090712
ENSDART00000127962 |
stox1
|
storkhead box 1 |
| chr16_-_12496632 | 2.68 |
ENSDART00000019941
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr9_-_10068004 | 2.61 |
ENSDART00000011922
ENSDART00000162818 |
spopla
|
speckle-type POZ protein-like a |
| chr12_+_27156943 | 2.61 |
ENSDART00000153030
ENSDART00000001737 |
skap1
|
src kinase associated phosphoprotein 1 |
| chr20_-_22193190 | 2.60 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr15_+_1004680 | 2.60 |
ENSDART00000157310
|
si:dkey-77f5.8
|
si:dkey-77f5.8 |
| chr13_+_33268657 | 2.51 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfcp2+tfcp2l1+ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.5 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 6.5 | 19.4 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 5.7 | 22.7 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 4.1 | 12.3 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 3.2 | 9.7 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 3.1 | 9.3 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 2.1 | 8.4 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 2.0 | 10.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 2.0 | 15.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 1.8 | 5.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.7 | 6.6 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 1.5 | 9.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 1.4 | 10.0 | GO:0070285 | pigment cell development(GO:0070285) |
| 1.3 | 4.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 1.3 | 10.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 1.2 | 4.7 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 1.2 | 10.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 1.1 | 14.7 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 1.1 | 7.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 1.0 | 4.1 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 1.0 | 5.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 1.0 | 9.0 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 1.0 | 19.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.9 | 2.8 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.9 | 2.6 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.8 | 12.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.8 | 4.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.7 | 4.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.7 | 23.2 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.7 | 4.7 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.6 | 5.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.6 | 3.4 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.6 | 12.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.5 | 3.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.5 | 18.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.5 | 1.6 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.5 | 2.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.5 | 3.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.5 | 4.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.5 | 2.5 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.5 | 8.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.5 | 10.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.5 | 3.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.5 | 13.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.5 | 2.3 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.5 | 10.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.4 | 5.2 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.4 | 11.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.4 | 8.0 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.4 | 2.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.4 | 15.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 3.4 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.4 | 11.7 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.4 | 2.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.4 | 6.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.4 | 2.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 3.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.3 | 1.0 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.3 | 3.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.3 | 3.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.3 | 3.0 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.3 | 1.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 3.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.3 | 1.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.3 | 8.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 1.9 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.3 | 1.3 | GO:2000301 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.3 | 2.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 31.6 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.3 | 15.9 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.2 | 6.1 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.2 | 2.7 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 | 3.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 0.9 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 1.8 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.2 | 3.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 3.5 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.2 | 6.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 5.9 | GO:0090148 | membrane fission(GO:0090148) |
| 0.2 | 2.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 2.6 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.2 | 17.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.2 | 2.9 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.2 | 0.6 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.1 | 18.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 4.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 2.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 2.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.9 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 7.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 1.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 9.3 | GO:0050673 | epithelial cell proliferation(GO:0050673) |
| 0.1 | 3.8 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 1.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.6 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 3.9 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 0.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 14.3 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.1 | 1.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 2.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 8.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 2.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.3 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.1 | 2.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.5 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 17.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.7 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 3.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.6 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.2 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 1.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 1.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 3.5 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 2.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 1.0 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 2.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 3.9 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 4.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.4 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 2.0 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 4.4 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 1.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 13.4 | GO:0006325 | chromatin organization(GO:0006325) |
| 0.0 | 1.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 3.6 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 2.5 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 2.9 | GO:0072594 | establishment of protein localization to organelle(GO:0072594) |
| 0.0 | 6.2 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 1.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.4 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 19.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 4.3 | 12.9 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 2.9 | 11.7 | GO:0061702 | inflammasome complex(GO:0061702) |
| 2.2 | 17.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.8 | 5.4 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 1.4 | 21.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 1.0 | 3.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.7 | 14.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.6 | 3.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.5 | 6.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.5 | 2.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.5 | 2.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 2.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 2.8 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.4 | 6.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 1.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 1.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.3 | 41.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.3 | 3.3 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.2 | 2.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 11.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 28.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 4.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 2.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 11.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 1.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 4.6 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 5.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 5.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 5.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 29.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 6.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 9.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 2.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 7.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.3 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 9.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 2.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 4.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 5.9 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 1.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 17.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 17.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 20.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 8.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 4.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 37.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 3.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 16.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 5.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 3.9 | 15.8 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 2.5 | 10.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 2.1 | 10.7 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 2.0 | 10.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 1.9 | 19.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.6 | 14.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 1.6 | 9.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 1.3 | 5.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.2 | 4.7 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 1.2 | 10.5 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 1.1 | 7.5 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 1.0 | 3.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.9 | 20.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.8 | 7.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.8 | 11.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.8 | 3.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.8 | 21.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.8 | 4.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.7 | 4.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.7 | 7.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.7 | 10.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.7 | 2.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.6 | 8.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.6 | 2.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.6 | 11.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.6 | 3.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 3.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 2.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.4 | 8.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.4 | 2.6 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.4 | 3.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 2.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.4 | 2.9 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 1.6 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.4 | 8.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.4 | 9.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.4 | 47.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 2.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.4 | 6.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.3 | 7.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 5.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 7.2 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.3 | 10.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 2.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 41.6 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.3 | 2.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 0.8 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.3 | 3.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 25.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 3.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 0.9 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 2.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 6.6 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.2 | 2.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 0.9 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 0.7 | GO:0046978 | MHC class I protein binding(GO:0042288) TAP1 binding(GO:0046978) |
| 0.2 | 1.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 1.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 8.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 5.2 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.2 | 4.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 2.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.2 | 3.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 3.8 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 14.3 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 2.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 7.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 2.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 3.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 19.4 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 0.6 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.1 | 0.3 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 2.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 2.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 7.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 2.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 3.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 3.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 4.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 2.6 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 41.9 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 3.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 5.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 8.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 4.7 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 10.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 3.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 8.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 5.4 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
| 0.0 | 4.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 3.1 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 14.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 15.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 2.9 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.3 | 4.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 7.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 10.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 4.2 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 19.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 7.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.9 | 14.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.7 | 10.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 4.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 4.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 15.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 2.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 3.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 4.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 2.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 3.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 3.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 3.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 10.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 4.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 2.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.5 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 3.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |