Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
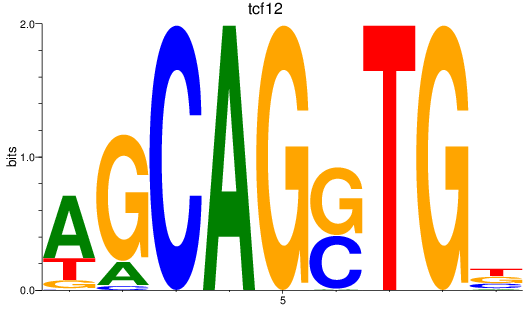
Results for tcf12
Z-value: 2.32
Transcription factors associated with tcf12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf12
|
ENSDARG00000004714 | transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf12 | dr11_v1_chr7_-_52709759_52709935 | 0.60 | 1.1e-10 | Click! |
Activity profile of tcf12 motif
Sorted Z-values of tcf12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_36911118 | 38.27 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr15_-_15357178 | 37.59 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr4_-_14328997 | 35.41 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr19_+_30633453 | 35.30 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr10_+_15777258 | 32.85 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr2_-_16565690 | 32.55 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr16_-_17207754 | 30.27 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr10_+_15777064 | 29.97 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr20_+_27020201 | 28.91 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr11_+_7324704 | 26.79 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr25_-_19433244 | 26.53 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr19_+_233143 | 26.30 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr23_+_28648864 | 25.51 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr3_-_28075756 | 25.36 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr10_-_17745345 | 24.87 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr21_-_19018455 | 24.83 |
ENSDART00000080256
|
nefma
|
neurofilament, medium polypeptide a |
| chr9_+_34425736 | 24.46 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr11_-_101758 | 23.46 |
ENSDART00000173015
|
elmo2
|
engulfment and cell motility 2 |
| chr1_+_59154521 | 23.33 |
ENSDART00000130089
ENSDART00000152456 |
soul5l
|
heme-binding protein soul5, like |
| chr14_-_33936524 | 22.89 |
ENSDART00000112438
|
si:ch73-335m24.5
|
si:ch73-335m24.5 |
| chr7_+_26224211 | 22.63 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr11_+_30057762 | 22.32 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr16_+_39159752 | 22.30 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr1_-_21409877 | 22.07 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr3_-_49566364 | 21.82 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr6_-_13308813 | 21.66 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr1_+_8662530 | 21.57 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr17_+_23298928 | 21.14 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr20_+_41549200 | 20.58 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr13_-_9318891 | 20.44 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr6_+_27667359 | 20.39 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr22_+_5106751 | 20.16 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr20_+_18580176 | 19.97 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr5_-_55395964 | 19.75 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr21_+_11468934 | 19.70 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr19_-_47526737 | 19.63 |
ENSDART00000186636
|
scg5
|
secretogranin V |
| chr16_-_36798783 | 19.38 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr14_-_21618005 | 19.31 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr13_+_40019001 | 19.26 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr5_+_3501859 | 19.09 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr5_+_42467867 | 19.09 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr1_-_38816685 | 18.71 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr17_-_26721007 | 18.69 |
ENSDART00000034580
|
calm1a
|
calmodulin 1a |
| chr2_-_36933472 | 18.33 |
ENSDART00000170405
|
BX470229.2
|
|
| chr17_-_16069905 | 18.32 |
ENSDART00000110383
|
map7a
|
microtubule-associated protein 7a |
| chr13_+_27314795 | 18.31 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr2_+_34967022 | 17.92 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr14_+_22172047 | 17.88 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr6_+_36942966 | 17.63 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr12_-_28881638 | 17.62 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr13_+_30054996 | 17.39 |
ENSDART00000110061
ENSDART00000186045 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr3_-_46818001 | 17.38 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr13_-_21688176 | 17.28 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr14_+_35748385 | 17.13 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr23_-_21453614 | 17.07 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr14_+_35748206 | 16.99 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr8_-_51404806 | 16.86 |
ENSDART00000060625
|
lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr8_-_4618653 | 16.72 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr1_-_25911292 | 16.71 |
ENSDART00000145012
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr8_+_24861264 | 16.67 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr6_-_35472923 | 16.60 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr2_+_34967210 | 16.58 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr18_-_8312848 | 16.52 |
ENSDART00000092033
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr19_-_5254699 | 16.52 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr17_-_751083 | 16.34 |
ENSDART00000188606
|
CABZ01102109.1
|
|
| chr16_+_43152727 | 16.27 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr11_+_37144328 | 16.22 |
ENSDART00000162830
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr5_-_46896541 | 16.13 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr5_+_22098591 | 15.89 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr5_-_10768258 | 15.89 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr13_+_4405282 | 15.63 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr7_+_50053339 | 15.63 |
ENSDART00000174308
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr5_-_30088070 | 15.57 |
ENSDART00000078115
ENSDART00000134209 |
sdhda
|
succinate dehydrogenase complex, subunit D, integral membrane protein a |
| chr16_+_7626535 | 15.52 |
ENSDART00000182670
ENSDART00000065514 ENSDART00000150212 |
stx12l
|
syntaxin 12, like |
| chr23_-_30781875 | 15.49 |
ENSDART00000114628
ENSDART00000180949 ENSDART00000191313 |
myt1a
|
myelin transcription factor 1a |
| chr3_-_46817838 | 15.46 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr9_+_4609746 | 15.43 |
ENSDART00000075420
|
rprma
|
reprimo, TP53 dependent G2 arrest mediator candidate a |
| chr4_+_3482312 | 15.36 |
ENSDART00000109044
|
grm8a
|
glutamate receptor, metabotropic 8a |
| chr3_-_36115339 | 15.36 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr5_-_38384289 | 15.36 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr5_+_23118470 | 15.21 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr24_-_24146875 | 14.93 |
ENSDART00000173052
|
map7d2b
|
MAP7 domain containing 2b |
| chr19_-_7450796 | 14.75 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr19_-_6385594 | 14.70 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr9_-_32753535 | 14.66 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr8_-_53198154 | 14.66 |
ENSDART00000083416
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr16_-_12984631 | 14.65 |
ENSDART00000184863
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr20_-_34801181 | 14.64 |
ENSDART00000048375
ENSDART00000132426 |
stmn4
|
stathmin-like 4 |
| chr16_+_4055331 | 14.55 |
ENSDART00000128978
|
CR391998.1
|
|
| chr13_+_51579851 | 14.53 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr22_+_5103349 | 14.53 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr12_-_10220036 | 14.48 |
ENSDART00000134619
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr13_+_11439486 | 14.44 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr3_-_38692920 | 14.29 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr6_+_59967994 | 14.25 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr19_+_8144556 | 14.23 |
ENSDART00000027274
ENSDART00000147218 |
efna3a
|
ephrin-A3a |
| chr13_+_36633355 | 14.18 |
ENSDART00000135612
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr10_-_41450367 | 14.15 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr17_+_23937262 | 14.11 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr8_-_7232413 | 13.84 |
ENSDART00000092426
|
grip2a
|
glutamate receptor interacting protein 2a |
| chr21_+_11468642 | 13.80 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr19_-_9711472 | 13.70 |
ENSDART00000016197
ENSDART00000175075 |
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr3_-_21061931 | 13.65 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr8_-_4596662 | 13.58 |
ENSDART00000138199
|
sept5a
|
septin 5a |
| chr14_+_11103718 | 13.56 |
ENSDART00000161311
|
nexmifb
|
neurite extension and migration factor b |
| chr14_+_21783229 | 13.46 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr13_+_30055171 | 13.44 |
ENSDART00000143581
ENSDART00000132027 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr6_-_16667886 | 13.37 |
ENSDART00000180854
ENSDART00000190116 |
unc80
|
unc-80 homolog (C. elegans) |
| chr5_-_55395384 | 13.35 |
ENSDART00000147298
ENSDART00000082577 |
prune2
|
prune homolog 2 (Drosophila) |
| chr19_-_30565122 | 13.33 |
ENSDART00000185650
|
hpcal4
|
hippocalcin like 4 |
| chr13_+_24834199 | 13.32 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr23_-_3674443 | 13.31 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr11_-_1291012 | 13.30 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr21_+_7823146 | 13.22 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr19_-_13774502 | 13.12 |
ENSDART00000159711
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr13_+_17702853 | 13.07 |
ENSDART00000145438
|
si:dkey-27m7.4
|
si:dkey-27m7.4 |
| chr5_-_13766651 | 12.97 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr16_+_45456066 | 12.85 |
ENSDART00000093365
|
syngap1b
|
synaptic Ras GTPase activating protein 1b |
| chr1_+_32521469 | 12.82 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr15_-_47937204 | 12.80 |
ENSDART00000154705
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr25_-_8030425 | 12.78 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr24_-_20641000 | 12.72 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr23_+_6795531 | 12.53 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr11_+_30058139 | 12.53 |
ENSDART00000112254
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr11_-_30352333 | 12.50 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr14_-_32258759 | 12.49 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr23_+_19594608 | 12.41 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr4_+_11140197 | 12.40 |
ENSDART00000067264
|
cracr2ab
|
calcium release activated channel regulator 2Ab |
| chr6_+_55277419 | 12.35 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr14_+_14225048 | 12.34 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr21_-_41305748 | 12.33 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr11_+_30000814 | 12.30 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr14_-_30490763 | 12.27 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr15_+_28482862 | 12.27 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr14_-_27121854 | 12.17 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr12_+_5189776 | 12.13 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr5_+_28398449 | 12.09 |
ENSDART00000165292
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr3_-_62380146 | 12.07 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr23_+_6272638 | 12.04 |
ENSDART00000190366
|
syt2a
|
synaptotagmin IIa |
| chr20_+_88168 | 12.03 |
ENSDART00000149283
|
zgc:112001
|
zgc:112001 |
| chr5_+_20030414 | 11.96 |
ENSDART00000181430
ENSDART00000047841 ENSDART00000182813 |
sgsm1a
|
small G protein signaling modulator 1a |
| chr22_+_10215558 | 11.82 |
ENSDART00000063274
|
kctd6a
|
potassium channel tetramerization domain containing 6a |
| chr6_-_38418862 | 11.77 |
ENSDART00000104135
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr7_-_58130703 | 11.72 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr4_+_12617108 | 11.70 |
ENSDART00000134362
ENSDART00000112860 |
lmo3
|
LIM domain only 3 |
| chr6_-_20875111 | 11.66 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr19_+_32979331 | 11.65 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr9_+_38983895 | 11.58 |
ENSDART00000144893
|
map2
|
microtubule-associated protein 2 |
| chr23_+_21455152 | 11.53 |
ENSDART00000158511
ENSDART00000161321 ENSDART00000160731 ENSDART00000137573 |
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr7_-_23996133 | 11.51 |
ENSDART00000173761
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr11_+_36180349 | 11.49 |
ENSDART00000012940
|
grm2b
|
glutamate receptor, metabotropic 2b |
| chr2_+_22702488 | 11.47 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr23_-_39849155 | 11.45 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr23_-_21463788 | 11.43 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr2_+_38881165 | 11.43 |
ENSDART00000141850
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr9_-_27648683 | 11.40 |
ENSDART00000017292
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr8_-_43158486 | 11.32 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr24_-_38384432 | 11.22 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr21_+_25935845 | 11.22 |
ENSDART00000141149
|
caln2
|
calneuron 2 |
| chr2_-_31735142 | 11.17 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr25_-_12203952 | 11.16 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr5_+_15667874 | 11.15 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr5_-_23280098 | 11.12 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr12_-_29624638 | 11.12 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr7_+_29951997 | 11.09 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr15_-_26636826 | 11.08 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr15_+_47161917 | 11.02 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr10_-_24343507 | 10.99 |
ENSDART00000002974
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr2_+_22694382 | 10.92 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr14_-_23737674 | 10.92 |
ENSDART00000005842
|
fgf1a
|
fibroblast growth factor 1a |
| chr14_+_45925810 | 10.90 |
ENSDART00000189543
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr3_-_22191132 | 10.89 |
ENSDART00000154226
ENSDART00000155528 ENSDART00000155190 |
maptb
|
microtubule-associated protein tau b |
| chr11_+_30161168 | 10.85 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr21_-_43952958 | 10.79 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr23_-_18057851 | 10.75 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr20_-_37629084 | 10.75 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr3_+_36515376 | 10.72 |
ENSDART00000161652
|
si:dkeyp-72e1.9
|
si:dkeyp-72e1.9 |
| chr10_+_8680730 | 10.71 |
ENSDART00000011987
|
isl1l
|
islet1, like |
| chr1_-_14233815 | 10.67 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr9_-_24050257 | 10.65 |
ENSDART00000090414
|
ackr3a
|
atypical chemokine receptor 3a |
| chr5_+_20148671 | 10.64 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr25_+_35683956 | 10.53 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr14_+_21783400 | 10.48 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr18_-_5692292 | 10.47 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr14_+_24215046 | 10.43 |
ENSDART00000079215
|
stc2a
|
stanniocalcin 2a |
| chr16_+_46148990 | 10.37 |
ENSDART00000083919
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr5_+_42402536 | 10.36 |
ENSDART00000186754
|
BX548073.11
|
|
| chr17_+_23938283 | 10.35 |
ENSDART00000184391
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr13_+_23157053 | 10.34 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr23_-_5032587 | 10.29 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr15_+_16908085 | 10.25 |
ENSDART00000186870
|
ypel2b
|
yippee-like 2b |
| chr9_-_43375205 | 10.23 |
ENSDART00000138436
|
znf385b
|
zinc finger protein 385B |
| chr2_-_33676494 | 10.17 |
ENSDART00000141192
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr17_-_19019635 | 10.15 |
ENSDART00000126666
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_-_10320676 | 10.13 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr11_+_1845787 | 10.07 |
ENSDART00000173062
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr2_+_26237322 | 10.04 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr13_-_41155472 | 10.02 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr6_-_14139503 | 9.99 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr3_+_46724528 | 9.96 |
ENSDART00000181358
|
pde4a
|
phosphodiesterase 4A, cAMP-specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.0 | 42.1 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 7.2 | 28.8 | GO:2000048 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 6.2 | 67.8 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 5.6 | 22.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 5.6 | 16.7 | GO:0015824 | proline transport(GO:0015824) |
| 5.4 | 26.8 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 5.2 | 5.2 | GO:0042478 | regulation of eye photoreceptor cell development(GO:0042478) |
| 4.8 | 19.4 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 4.3 | 17.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 4.1 | 16.6 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 4.1 | 24.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 3.6 | 10.9 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 3.4 | 10.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 3.3 | 16.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 3.3 | 16.3 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 3.1 | 71.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 3.1 | 15.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 3.1 | 9.3 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 3.0 | 9.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 3.0 | 9.0 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 3.0 | 14.8 | GO:0090200 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 2.7 | 8.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 2.6 | 46.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 2.4 | 54.9 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 2.3 | 11.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 2.2 | 40.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 2.2 | 8.8 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 2.2 | 41.3 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 2.2 | 4.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 2.1 | 8.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 2.1 | 28.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 2.0 | 8.2 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 2.0 | 6.0 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 2.0 | 9.9 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 1.9 | 7.8 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 1.9 | 19.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 1.7 | 6.9 | GO:0015677 | copper ion import(GO:0015677) |
| 1.7 | 22.1 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 1.7 | 6.7 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 1.7 | 18.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 1.7 | 5.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 1.6 | 6.6 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 1.6 | 8.0 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 1.6 | 17.2 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 1.5 | 6.2 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 1.5 | 4.5 | GO:0016264 | gap junction assembly(GO:0016264) |
| 1.5 | 8.9 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 1.5 | 23.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 1.5 | 11.6 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 1.5 | 7.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 1.4 | 4.3 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 1.4 | 23.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 1.4 | 6.9 | GO:0034505 | tooth mineralization(GO:0034505) |
| 1.3 | 6.6 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 1.3 | 9.2 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 1.3 | 10.5 | GO:0021634 | optic nerve formation(GO:0021634) |
| 1.3 | 6.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 1.3 | 16.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 1.3 | 3.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.3 | 8.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 1.3 | 3.8 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 1.2 | 19.8 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 1.2 | 30.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 1.2 | 30.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 1.1 | 6.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.1 | 7.6 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 1.1 | 3.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 1.1 | 17.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 1.0 | 5.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 1.0 | 6.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 1.0 | 6.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 1.0 | 20.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 1.0 | 5.0 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 1.0 | 3.0 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 1.0 | 3.9 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 1.0 | 3.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 1.0 | 33.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 1.0 | 2.9 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 1.0 | 2.9 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 1.0 | 2.9 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.9 | 4.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.9 | 3.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.9 | 6.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.9 | 1.9 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.9 | 11.1 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.9 | 4.6 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.9 | 4.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.9 | 4.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.9 | 2.7 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.9 | 7.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.9 | 17.1 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.9 | 21.0 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.9 | 4.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.9 | 32.0 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.9 | 8.6 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.8 | 4.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.8 | 5.9 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.8 | 3.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.8 | 5.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.8 | 23.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.8 | 9.0 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.8 | 4.8 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.8 | 4.8 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.8 | 12.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.8 | 2.4 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.8 | 1.5 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.8 | 5.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.8 | 6.8 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.8 | 7.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.7 | 6.7 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.7 | 2.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.7 | 14.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.7 | 2.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.7 | 8.0 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.7 | 26.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.7 | 4.3 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.7 | 8.6 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.7 | 2.1 | GO:1903060 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.7 | 9.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.7 | 3.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.7 | 13.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.7 | 4.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 24.5 | GO:0001764 | neuron migration(GO:0001764) |
| 0.7 | 7.7 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.7 | 2.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.7 | 27.4 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.7 | 13.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.7 | 29.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.7 | 14.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.7 | 6.0 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.7 | 11.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.7 | 0.7 | GO:0061193 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.7 | 22.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.6 | 5.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.6 | 5.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.6 | 25.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.6 | 7.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.6 | 6.9 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.6 | 1.9 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.6 | 5.6 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.6 | 22.8 | GO:0010107 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.6 | 0.6 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.6 | 4.3 | GO:0019857 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.6 | 20.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.6 | 8.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.6 | 4.2 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.6 | 4.2 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.6 | 1.8 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.6 | 2.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.6 | 1.8 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) eurydendroid cell differentiation(GO:0021755) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.6 | 12.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.6 | 2.3 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.6 | 17.4 | GO:0048883 | neuromast primordium migration(GO:0048883) posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.6 | 5.8 | GO:0042044 | fluid transport(GO:0042044) |
| 0.6 | 6.3 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.6 | 4.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.6 | 7.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.6 | 3.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.5 | 1.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.5 | 23.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.5 | 5.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.5 | 4.8 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.5 | 3.7 | GO:0070207 | protein trimerization(GO:0070206) protein homotrimerization(GO:0070207) |
| 0.5 | 3.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 3.7 | GO:0044539 | very long-chain fatty acid catabolic process(GO:0042760) long-chain fatty acid import(GO:0044539) |
| 0.5 | 18.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.5 | 16.0 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.5 | 2.0 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.5 | 6.8 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.5 | 6.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.5 | 1.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.5 | 7.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.5 | 8.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.5 | 6.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.5 | 6.0 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.5 | 5.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.5 | 7.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.5 | 15.9 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.4 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 9.9 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.4 | 1.8 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.4 | 7.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.4 | 4.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.4 | 1.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 4.3 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.4 | 6.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 3.0 | GO:0030728 | ovulation(GO:0030728) |
| 0.4 | 2.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 45.5 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.4 | 9.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.4 | 5.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 2.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.4 | 1.6 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.4 | 0.4 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.4 | 3.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.4 | 2.7 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.4 | 22.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.4 | 2.7 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.4 | 16.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.4 | 4.9 | GO:0086001 | cardiac muscle cell action potential(GO:0086001) |
| 0.4 | 2.6 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.4 | 9.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.4 | 22.3 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.4 | 10.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.4 | 2.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.4 | 5.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 3.6 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.4 | 38.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.4 | 2.1 | GO:0043090 | amino acid import(GO:0043090) L-arginine import(GO:0043091) L-amino acid import(GO:0043092) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.4 | 19.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.3 | 1.4 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.3 | 17.8 | GO:0006821 | chloride transport(GO:0006821) |
| 0.3 | 2.4 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.3 | 18.0 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.3 | 18.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 15.4 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.3 | 1.4 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.3 | 12.8 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.3 | 4.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 3.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.3 | 2.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.3 | 3.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 2.5 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.3 | 0.6 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.3 | 1.6 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.3 | 2.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 12.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.3 | 28.1 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.3 | 9.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.3 | 24.3 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.3 | 22.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 0.9 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.3 | 8.5 | GO:0016358 | dendrite development(GO:0016358) |
| 0.3 | 2.0 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.3 | 2.8 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.3 | 1.1 | GO:0042727 | flavin-containing compound metabolic process(GO:0042726) flavin-containing compound biosynthetic process(GO:0042727) |
| 0.3 | 5.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.3 | 6.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.3 | 5.1 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.3 | 2.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 7.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 7.5 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.3 | 16.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.3 | 3.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 2.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 1.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.3 | 29.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.3 | 1.8 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 | 1.2 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.2 | 1.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 0.9 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 2.4 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 8.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.2 | 27.0 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 11.6 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.2 | 62.2 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.2 | 1.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 2.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 2.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 2.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 2.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 7.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 3.0 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 4.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.2 | 4.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 2.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.2 | 5.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 1.0 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 2.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.2 | 2.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 4.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 3.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.2 | 2.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 2.9 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.2 | 3.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 1.1 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.2 | 1.9 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 7.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.2 | 3.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 5.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.2 | 2.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 4.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.2 | 11.7 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.2 | 8.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 6.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.2 | 5.4 | GO:0060914 | heart formation(GO:0060914) |
| 0.2 | 3.1 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.2 | 0.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 0.8 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 8.3 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.2 | 11.1 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.2 | 5.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 2.0 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.2 | 2.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.2 | 1.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 1.3 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 3.1 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 4.9 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 11.0 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 1.1 | GO:0046958 | learning(GO:0007612) nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 4.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 15.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 6.0 | GO:0044782 | cilium organization(GO:0044782) |
| 0.1 | 3.5 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 2.2 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.1 | 24.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.0 | GO:0003401 | axis elongation(GO:0003401) |
| 0.1 | 1.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.9 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.6 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 3.4 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 25.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.4 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.5 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 17.3 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.1 | 1.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.7 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.5 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 5.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.4 | GO:0010259 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 10.0 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 6.9 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 0.8 | GO:0032412 | regulation of ion transmembrane transporter activity(GO:0032412) |
| 0.1 | 2.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 3.4 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 1.6 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.1 | 0.9 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 9.7 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.1 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.0 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.1 | 0.7 | GO:0045923 | negative regulation of cholesterol storage(GO:0010887) positive regulation of fatty acid metabolic process(GO:0045923) |
| 0.1 | 3.8 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 1.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 19.0 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.1 | 0.9 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 2.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 7.3 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 7.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 19.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 7.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 2.0 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 2.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 1.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 3.7 | GO:0016573 | internal protein amino acid acetylation(GO:0006475) histone acetylation(GO:0016573) internal peptidyl-lysine acetylation(GO:0018393) |
| 0.0 | 1.3 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 1.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 1.1 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.5 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.2 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.2 | GO:0006448 | regulation of translational elongation(GO:0006448) |
| 0.0 | 3.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 7.9 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 10.1 | GO:0055085 | transmembrane transport(GO:0055085) |
| 0.0 | 0.8 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.3 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.2 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 5.0 | 24.8 | GO:0005883 | neurofilament(GO:0005883) |
| 4.2 | 42.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 3.1 | 15.6 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 2.8 | 28.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 2.8 | 42.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 2.8 | 11.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 2.6 | 15.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 2.4 | 28.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 2.1 | 27.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 2.1 | 22.8 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 2.0 | 14.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 2.0 | 12.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 1.8 | 80.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 1.7 | 11.7 | GO:0030315 | T-tubule(GO:0030315) |
| 1.6 | 21.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.5 | 4.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 1.4 | 21.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 1.4 | 8.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.4 | 11.1 | GO:0016586 | RSC complex(GO:0016586) |
| 1.3 | 32.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 1.2 | 21.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 1.2 | 11.6 | GO:0070449 | elongin complex(GO:0070449) |
| 1.1 | 20.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 1.1 | 5.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 1.1 | 40.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 1.0 | 5.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 1.0 | 34.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.0 | 3.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 1.0 | 41.5 | GO:0030426 | growth cone(GO:0030426) |
| 1.0 | 32.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.9 | 3.5 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.8 | 6.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.8 | 4.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.7 | 5.9 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.7 | 12.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.7 | 16.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.7 | 5.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.7 | 4.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.7 | 11.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.7 | 52.2 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.7 | 41.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.6 | 53.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.6 | 1.8 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.6 | 20.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.6 | 3.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.6 | 4.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.6 | 13.3 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.6 | 41.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.6 | 4.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.6 | 15.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.6 | 96.5 | GO:0030424 | axon(GO:0030424) |
| 0.5 | 8.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 20.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.5 | 9.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.4 | 23.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.4 | 3.5 | GO:0030666 | endocytic vesicle membrane(GO:0030666) phagocytic vesicle membrane(GO:0030670) |
| 0.4 | 7.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.4 | 45.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.4 | 6.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.4 | 100.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.4 | 5.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 1.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 6.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 2.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 3.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 3.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 5.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.3 | 2.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 14.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 2.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 24.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.3 | 27.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.3 | 33.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.3 | 19.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.3 | 17.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 9.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.2 | 15.2 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.2 | 27.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 3.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 4.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 1.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.2 | GO:0097223 | sperm part(GO:0097223) |
| 0.2 | 4.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 1.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 1.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 3.8 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.2 | 1.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 1.9 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 5.1 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 16.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 28.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.2 | 0.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 6.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 2.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 10.5 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.1 | 2.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 4.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 2.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 4.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.4 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 0.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 29.2 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 1.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 63.1 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 4.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 4.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 8.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 3.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 19.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.1 | 75.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.8 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.0 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.9 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 6.2 | 67.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 5.1 | 25.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 4.1 | 12.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 3.7 | 56.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 3.6 | 32.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 3.3 | 9.9 | GO:0009013 | succinate-semialdehyde dehydrogenase (NAD+) activity(GO:0004777) succinate-semialdehyde dehydrogenase [NAD(P)+] activity(GO:0009013) |
| 3.0 | 21.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 2.8 | 16.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 2.7 | 10.9 | GO:0044548 | S100 protein binding(GO:0044548) |
| 2.6 | 62.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 2.4 | 21.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 2.2 | 20.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 2.1 | 42.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 2.1 | 6.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 2.0 | 12.0 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 2.0 | 35.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 1.9 | 5.8 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 1.9 | 17.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.8 | 7.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.7 | 15.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.7 | 6.9 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 1.6 | 11.5 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 1.6 | 8.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 1.6 | 17.9 | GO:0008556 | potassium-transporting ATPase activity(GO:0008556) |
| 1.6 | 34.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 1.6 | 11.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.5 | 4.5 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 1.5 | 7.3 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 1.5 | 7.3 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 1.4 | 4.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.4 | 12.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.4 | 28.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 1.4 | 9.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 1.3 | 6.6 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 1.3 | 18.4 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 1.3 | 3.9 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 1.3 | 11.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.2 | 5.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 1.2 | 45.9 | GO:0019003 | GDP binding(GO:0019003) |
| 1.2 | 19.8 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 1.2 | 17.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 1.2 | 8.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 1.2 | 4.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.1 | 9.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.1 | 6.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 1.1 | 6.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 1.1 | 7.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 1.1 | 28.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.0 | 6.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.0 | 8.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.0 | 29.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.9 | 8.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.9 | 44.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.9 | 11.0 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.9 | 16.9 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.9 | 2.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.8 | 5.9 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.8 | 4.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.8 | 6.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.8 | 3.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.8 | 15.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.8 | 11.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.8 | 7.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.8 | 15.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.8 | 10.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.8 | 15.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.8 | 3.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.8 | 3.8 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.7 | 3.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.7 | 11.9 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.7 | 11.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.7 | 9.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.7 | 2.2 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.7 | 11.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.7 | 8.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.7 | 2.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.7 | 15.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.7 | 10.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.7 | 5.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.7 | 11.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.7 | 32.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.7 | 6.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.6 | 5.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.6 | 2.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.6 | 16.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.6 | 12.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.6 | 4.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.6 | 20.3 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.6 | 27.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.6 | 3.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 1.8 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.6 | 9.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.6 | 4.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 10.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.6 | 29.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.6 | 2.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.6 | 13.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.5 | 6.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.5 | 15.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.5 | 12.5 | GO:0015298 | solute:cation antiporter activity(GO:0015298) |
| 0.5 | 3.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 5.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 7.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.5 | 7.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.5 | 4.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.5 | 2.5 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.5 | 6.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.5 | 9.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.5 | 12.0 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.5 | 16.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 6.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.5 | 13.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 16.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 1.9 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.4 | 7.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 144.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.4 | 28.0 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.4 | 2.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 9.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 2.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 9.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 2.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 3.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 5.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 6.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 5.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.4 | 1.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.4 | 1.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 4.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.4 | 2.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 8.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 2.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 4.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.3 | 2.4 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.3 | 23.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.3 | 8.0 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.3 | 3.5 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.3 | 117.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 1.6 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.3 | 2.5 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.3 | 15.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 19.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.3 | 3.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.3 | 3.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 2.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.3 | 16.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 2.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 7.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.3 | 2.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 2.4 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 5.7 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.2 | 47.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.2 | 1.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 10.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.2 | 0.7 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 2.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 31.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.2 | 0.9 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 6.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 84.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 1.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 5.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 3.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 7.5 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 0.9 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.2 | 3.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 17.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 9.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 2.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 4.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.2 | 8.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 5.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 9.7 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.2 | 12.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 7.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 11.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.2 | 11.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 10.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 13.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 3.9 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.2 | 0.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 3.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 6.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.8 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 4.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 11.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 9.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.0 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 6.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 2.4 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 4.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 2.9 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 4.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 5.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 5.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 5.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.3 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 0.3 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 0.9 | GO:0017136 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.1 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 3.3 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 3.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 3.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 5.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 2.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 3.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.5 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.1 | 1.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 3.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 0.5 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 4.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 2.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.4 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 3.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.5 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 1.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 9.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 5.0 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.9 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 15.7 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.8 | 16.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.8 | 4.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.8 | 10.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.8 | 32.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.6 | 19.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.6 | 18.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.6 | 8.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.5 | 8.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.4 | 7.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 3.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.4 | 9.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.4 | 4.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.4 | 11.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 14.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.3 | 5.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 5.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 4.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 2.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 6.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 1.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 5.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 3.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 3.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 3.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 10.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.6 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 4.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 8.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 2.5 | 37.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.2 | 16.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 1.2 | 16.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 1.0 | 4.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.9 | 17.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.8 | 11.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.7 | 4.4 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.7 | 7.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.7 | 8.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.7 | 7.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.6 | 5.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.6 | 6.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.6 | 6.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.6 | 13.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.6 | 8.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.5 | 10.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.5 | 8.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.5 | 7.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 4.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 11.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.4 | 3.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 9.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.4 | 2.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.4 | 3.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 3.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.3 | 6.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 26.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 3.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 1.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 5.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 4.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 8.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 5.9 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.2 | 2.8 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 13.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 7.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 21.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.9 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 3.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.0 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 0.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |