Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
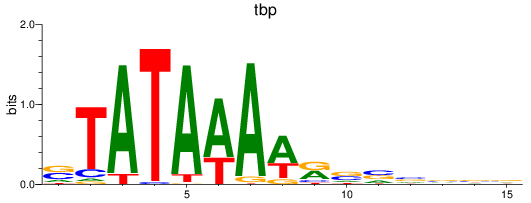
Results for tbp
Z-value: 2.76
Transcription factors associated with tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbp
|
ENSDARG00000014994 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbp | dr11_v1_chr13_-_24396199_24396199 | -0.80 | 1.0e-21 | Click! |
Activity profile of tbp motif
Sorted Z-values of tbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_32818607 | 46.64 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr21_+_20383837 | 42.96 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr5_-_36837846 | 42.10 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr1_+_55137943 | 40.40 |
ENSDART00000138070
ENSDART00000150510 ENSDART00000133472 ENSDART00000136378 |
mb
|
myoglobin |
| chr23_+_45456490 | 37.20 |
ENSDART00000036631
|
cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr5_-_32274383 | 34.72 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr25_+_35020529 | 34.29 |
ENSDART00000158016
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr12_-_17712393 | 33.94 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr9_+_23900703 | 29.65 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr22_-_263117 | 29.15 |
ENSDART00000158134
|
zgc:66156
|
zgc:66156 |
| chr22_-_282498 | 28.24 |
ENSDART00000182766
|
CABZ01079178.1
|
|
| chr5_-_71722257 | 26.92 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr15_-_5815006 | 26.54 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr16_+_1471 | 26.31 |
ENSDART00000181972
|
CABZ01090785.1
|
|
| chr8_+_48613040 | 25.83 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr23_-_35082494 | 25.15 |
ENSDART00000189809
|
BX294434.1
|
|
| chr7_-_16562200 | 23.97 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr20_-_9980318 | 23.61 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr5_-_64168415 | 21.88 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr24_-_25691020 | 21.54 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr13_-_12660318 | 20.95 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr23_+_27675581 | 20.57 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr8_-_24970790 | 20.24 |
ENSDART00000141267
|
si:ch211-199o1.2
|
si:ch211-199o1.2 |
| chr2_+_24507597 | 19.69 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr1_-_49225890 | 19.27 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr22_-_294700 | 19.11 |
ENSDART00000189179
|
CABZ01079179.1
|
|
| chr23_+_25305431 | 18.54 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr8_+_48609521 | 17.87 |
ENSDART00000060765
|
nppb
|
natriuretic peptide B |
| chr2_+_16780643 | 17.76 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr11_+_6456146 | 17.69 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr2_+_24507770 | 17.59 |
ENSDART00000154802
ENSDART00000052063 |
rps28
|
ribosomal protein S28 |
| chr13_-_2215213 | 17.59 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr6_-_54180699 | 17.53 |
ENSDART00000045901
|
rps10
|
ribosomal protein S10 |
| chr18_-_46010 | 17.49 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr2_+_15100742 | 16.38 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr2_+_16781015 | 15.60 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr6_+_269204 | 15.56 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr7_-_31441420 | 15.49 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr7_+_29951997 | 14.55 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr5_-_25583125 | 14.52 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr5_-_33259079 | 14.30 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr8_-_31369161 | 13.22 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr18_-_5527050 | 12.93 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr2_-_30182353 | 12.92 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr20_-_26846028 | 12.81 |
ENSDART00000136687
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr13_-_15702672 | 12.81 |
ENSDART00000144445
ENSDART00000168950 |
ckba
|
creatine kinase, brain a |
| chr14_-_33334065 | 12.40 |
ENSDART00000052761
|
rpl39
|
ribosomal protein L39 |
| chr16_+_53455638 | 12.05 |
ENSDART00000045792
ENSDART00000154189 |
rbm24b
|
RNA binding motif protein 24b |
| chr8_+_26818446 | 12.04 |
ENSDART00000134987
ENSDART00000138835 |
si:ch211-156j16.1
|
si:ch211-156j16.1 |
| chr21_-_11646878 | 11.96 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr16_-_45917322 | 11.87 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr9_-_22232902 | 11.76 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr25_+_30298377 | 11.59 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr3_+_5575313 | 11.56 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr16_+_32736588 | 11.56 |
ENSDART00000075191
ENSDART00000168358 |
zgc:172323
|
zgc:172323 |
| chr5_-_7834582 | 11.52 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr21_-_28523548 | 11.47 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr5_+_37978501 | 11.46 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr2_+_55365727 | 11.24 |
ENSDART00000162943
|
FP245456.1
|
|
| chr4_+_842010 | 11.04 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr4_+_7817996 | 10.90 |
ENSDART00000166809
|
si:ch1073-67j19.1
|
si:ch1073-67j19.1 |
| chr24_-_10021341 | 10.85 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr23_+_18103080 | 10.79 |
ENSDART00000010270
|
mfsd4ab
|
major facilitator superfamily domain containing 4Ab |
| chr9_-_33081978 | 10.66 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr3_-_57425961 | 10.58 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr9_-_22240052 | 10.50 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr11_+_13207898 | 10.46 |
ENSDART00000060310
|
atp5f1b
|
ATP synthase F1 subunit beta |
| chr7_-_73851280 | 10.30 |
ENSDART00000190053
|
FP236812.3
|
|
| chr21_+_30043054 | 10.15 |
ENSDART00000065448
|
fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr3_+_26135502 | 10.12 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr15_+_28368823 | 10.08 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr5_-_15851953 | 10.04 |
ENSDART00000173101
|
si:dkey-1k23.3
|
si:dkey-1k23.3 |
| chr17_+_27456804 | 10.00 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr12_-_20665164 | 9.93 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr23_+_43950674 | 9.85 |
ENSDART00000167813
|
corin
|
corin, serine peptidase |
| chr5_-_30615901 | 9.75 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr24_-_9991153 | 9.60 |
ENSDART00000137794
ENSDART00000106252 ENSDART00000188309 ENSDART00000188266 ENSDART00000188660 ENSDART00000185713 ENSDART00000179773 |
zgc:152652
|
zgc:152652 |
| chr22_-_817479 | 9.56 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr24_+_3307857 | 9.54 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr1_-_43920371 | 9.52 |
ENSDART00000109283
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr12_+_20641102 | 9.43 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr21_+_25221940 | 9.42 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr2_+_24536762 | 9.42 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr14_+_22132388 | 9.36 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr24_-_9979342 | 9.34 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr24_+_21621654 | 9.26 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr3_-_24456451 | 9.26 |
ENSDART00000024480
ENSDART00000156814 |
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr10_-_40557210 | 9.23 |
ENSDART00000135297
|
taar18b
|
trace amine associated receptor 18b |
| chr16_-_31598771 | 9.05 |
ENSDART00000016386
|
tg
|
thyroglobulin |
| chr2_+_55665322 | 9.02 |
ENSDART00000183636
ENSDART00000183814 |
klf2b
|
Kruppel-like factor 2b |
| chr24_-_27419198 | 8.95 |
ENSDART00000141124
|
ccl34b.4
|
chemokine (C-C motif) ligand 34b, duplicate 4 |
| chr1_+_23408622 | 8.95 |
ENSDART00000140706
|
chrna9
|
cholinergic receptor, nicotinic, alpha 9 |
| chr7_-_73717082 | 8.69 |
ENSDART00000164301
ENSDART00000082625 |
BX664721.2
|
|
| chr18_-_50799510 | 8.61 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr9_-_33063083 | 8.60 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr9_+_23770666 | 8.58 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr6_+_52918537 | 8.55 |
ENSDART00000174229
|
or137-1
|
odorant receptor, family H, subfamily 137, member 1 |
| chr11_+_25477643 | 8.48 |
ENSDART00000065941
|
opn1lw1
|
opsin 1 (cone pigments), long-wave-sensitive, 1 |
| chr12_+_48340133 | 8.48 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr2_-_127945 | 8.42 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr22_+_16497670 | 8.40 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr13_-_12645584 | 8.31 |
ENSDART00000176216
|
adh8b
|
alcohol dehydrogenase 8b |
| chr16_-_25085327 | 8.24 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr22_+_5752257 | 8.24 |
ENSDART00000143052
|
cox14
|
COX14 cytochrome c oxidase assembly factor |
| chr21_+_5531138 | 8.12 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr1_-_43920576 | 8.12 |
ENSDART00000191914
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr10_+_17026870 | 8.11 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr4_+_25706037 | 8.08 |
ENSDART00000141133
|
lamb1b
|
laminin, beta 1b |
| chr20_-_52928541 | 8.07 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr7_+_35068036 | 7.69 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr7_-_73845736 | 7.67 |
ENSDART00000193414
|
zgc:173552
|
zgc:173552 |
| chr17_-_20118145 | 7.65 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr10_+_38593645 | 7.63 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr24_-_9985019 | 7.62 |
ENSDART00000193536
ENSDART00000189595 |
zgc:171977
|
zgc:171977 |
| chr22_+_635813 | 7.47 |
ENSDART00000179067
|
CU856139.1
|
|
| chr3_+_15505275 | 7.43 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr7_-_66137065 | 7.21 |
ENSDART00000145843
|
pth1b
|
parathyroid hormone 1b |
| chr13_-_20381485 | 7.17 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr23_+_19655301 | 7.16 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr3_-_23643751 | 7.15 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr11_+_3575963 | 7.13 |
ENSDART00000077305
|
si:dkey-33m11.8
|
si:dkey-33m11.8 |
| chr6_-_49173891 | 6.98 |
ENSDART00000132867
|
ngfb
|
nerve growth factor b (beta polypeptide) |
| chr7_-_73846995 | 6.97 |
ENSDART00000188079
|
FP236812.4
|
|
| chr9_-_22355391 | 6.97 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr6_-_40755909 | 6.91 |
ENSDART00000026257
|
myhb
|
myosin, heavy chain b |
| chr21_+_25777425 | 6.85 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr25_+_35132090 | 6.82 |
ENSDART00000154377
|
si:dkey-261m9.6
|
si:dkey-261m9.6 |
| chr9_-_33081781 | 6.82 |
ENSDART00000165748
|
zgc:172053
|
zgc:172053 |
| chr8_-_22965214 | 6.75 |
ENSDART00000148178
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr2_+_17181777 | 6.74 |
ENSDART00000112063
|
ptger4c
|
prostaglandin E receptor 4 (subtype EP4) c |
| chr11_+_42478184 | 6.71 |
ENSDART00000089963
|
zgc:110286
|
zgc:110286 |
| chr7_+_5960491 | 6.70 |
ENSDART00000145370
|
zgc:112234
|
zgc:112234 |
| chr23_-_35195908 | 6.66 |
ENSDART00000122429
|
klf15
|
Kruppel-like factor 15 |
| chr21_-_280769 | 6.65 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr3_-_40276057 | 6.59 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr1_-_59169815 | 6.58 |
ENSDART00000100163
|
wu:fk65c09
|
wu:fk65c09 |
| chr10_-_7785930 | 6.58 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr19_+_10845953 | 6.48 |
ENSDART00000157589
|
apoa4a
|
apolipoprotein A-IV a |
| chr9_+_1139378 | 6.47 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr19_-_42557416 | 6.45 |
ENSDART00000163217
ENSDART00000128278 ENSDART00000162304 ENSDART00000166556 |
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr15_-_18232712 | 6.34 |
ENSDART00000081199
|
wu:fj20b03
|
wu:fj20b03 |
| chr10_+_36334502 | 6.32 |
ENSDART00000099400
|
or106-1
|
odorant receptor, family G, subfamily 106, member 1 |
| chr5_+_4366431 | 6.26 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr4_-_19033805 | 6.24 |
ENSDART00000133203
|
lepb
|
leptin b |
| chr9_-_45602978 | 6.21 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr3_+_1037946 | 6.20 |
ENSDART00000167590
ENSDART00000011111 |
zgc:153921
zgc:153921
|
zgc:153921 zgc:153921 |
| chr11_-_45152702 | 6.19 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr6_-_40756138 | 6.19 |
ENSDART00000156295
|
myhb
|
myosin, heavy chain b |
| chr22_+_25774750 | 6.15 |
ENSDART00000174421
|
AL929192.1
|
|
| chr11_-_40030139 | 6.13 |
ENSDART00000021916
|
uts2b
|
urotensin 2, beta |
| chr9_-_22272181 | 6.11 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr4_+_306036 | 6.10 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr6_+_41181869 | 6.08 |
ENSDART00000002046
|
opn1mw1
|
opsin 1 (cone pigments), medium-wave-sensitive, 1 |
| chr15_+_714203 | 6.07 |
ENSDART00000153847
|
si:dkey-7i4.24
|
si:dkey-7i4.24 |
| chr17_+_37227936 | 6.05 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr5_+_68807170 | 5.98 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr25_-_12982193 | 5.97 |
ENSDART00000159617
|
ccl39.5
|
chemokine (C-C motif) ligand 39, duplicate 5 |
| chr13_+_40501455 | 5.95 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr15_+_15390882 | 5.93 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr9_-_33062891 | 5.89 |
ENSDART00000161182
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr9_-_22245572 | 5.89 |
ENSDART00000114943
|
crygm2d4
|
crystallin, gamma M2d4 |
| chr20_+_46255057 | 5.86 |
ENSDART00000100536
|
taar14i
|
trace amine associated receptor 14i |
| chr3_+_52145511 | 5.85 |
ENSDART00000078485
|
adgre14
|
adhesion G protein-coupled receptor E14 |
| chr12_-_36268723 | 5.85 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr11_-_24016761 | 5.85 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr25_+_8407892 | 5.83 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr3_-_6709938 | 5.82 |
ENSDART00000172196
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr15_-_5222924 | 5.82 |
ENSDART00000128924
|
or128-4
|
odorant receptor, family E, subfamily 128, member 4 |
| chr5_+_21181047 | 5.81 |
ENSDART00000088506
|
arhgap25
|
Rho GTPase activating protein 25 |
| chr7_-_7810348 | 5.80 |
ENSDART00000171984
|
cxcl19
|
chemokine (C-X-C motif) ligand 19 |
| chr25_-_375882 | 5.78 |
ENSDART00000165705
|
szl
|
sizzled |
| chr9_-_22281854 | 5.71 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr7_+_5906327 | 5.71 |
ENSDART00000173160
|
zgc:112234
|
zgc:112234 |
| chr3_-_55121125 | 5.64 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr16_-_28091597 | 5.63 |
ENSDART00000166595
|
CR626941.1
|
|
| chr16_+_31542645 | 5.59 |
ENSDART00000163724
|
SLA (1 of many)
|
Src like adaptor |
| chr7_+_5976613 | 5.57 |
ENSDART00000173105
|
si:dkey-23a13.21
|
si:dkey-23a13.21 |
| chr18_+_14342326 | 5.57 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr19_-_2707048 | 5.55 |
ENSDART00000166112
|
il6
|
interleukin 6 (interferon, beta 2) |
| chr6_+_49052741 | 5.55 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr22_-_8499778 | 5.55 |
ENSDART00000142219
|
si:ch73-27e22.4
|
si:ch73-27e22.4 |
| chr13_+_14976108 | 5.48 |
ENSDART00000011520
|
noto
|
notochord homeobox |
| chr12_-_20665865 | 5.47 |
ENSDART00000183922
|
gip
|
gastric inhibitory polypeptide |
| chr21_+_25226558 | 5.40 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr20_-_22464250 | 5.37 |
ENSDART00000165904
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr9_+_33207574 | 5.30 |
ENSDART00000055897
ENSDART00000166030 |
si:ch211-125e6.11
|
si:ch211-125e6.11 |
| chr24_-_26854032 | 5.27 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr8_+_999421 | 5.27 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr20_+_46268906 | 5.25 |
ENSDART00000113372
|
taar14e
|
trace amine associated receptor 14e |
| chr3_-_36606884 | 5.25 |
ENSDART00000172003
|
si:dkeyp-72e1.6
|
si:dkeyp-72e1.6 |
| chr9_+_22080122 | 5.22 |
ENSDART00000065956
ENSDART00000136014 |
crygm2e
|
crystallin, gamma M2e |
| chr14_-_11507211 | 5.18 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr21_+_21673516 | 5.17 |
ENSDART00000147601
|
or125-6
|
odorant receptor, family E, subfamily 125, member 6 |
| chr21_-_42876565 | 5.14 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr20_-_33976937 | 5.13 |
ENSDART00000136834
|
sele
|
selectin E |
| chr5_-_22027357 | 5.10 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr19_-_3193912 | 5.10 |
ENSDART00000133159
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr19_-_3193443 | 5.08 |
ENSDART00000179855
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr8_+_37111643 | 5.08 |
ENSDART00000061336
|
ren
|
renin |
| chr21_+_25354106 | 5.06 |
ENSDART00000133406
|
or132-4
|
odorant receptor, family H, subfamily 132, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.6 | 43.7 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 6.0 | 24.0 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 5.8 | 17.5 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 4.5 | 13.5 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 4.2 | 54.9 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 4.2 | 29.3 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 3.8 | 26.9 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 3.8 | 34.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 3.6 | 21.9 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 3.4 | 30.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 3.0 | 33.4 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 3.0 | 12.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 3.0 | 29.9 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 2.6 | 10.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 2.5 | 2.5 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 2.5 | 10.1 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 2.4 | 54.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 2.3 | 11.5 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 2.3 | 9.1 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 2.2 | 6.6 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 2.0 | 6.1 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 2.0 | 40.4 | GO:0014823 | response to activity(GO:0014823) |
| 1.9 | 40.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 1.8 | 5.5 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 1.8 | 5.5 | GO:0048321 | axial mesoderm formation(GO:0048320) axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) |
| 1.7 | 8.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 1.7 | 12.0 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 1.7 | 5.0 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 1.7 | 6.7 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 1.6 | 6.3 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 1.6 | 4.7 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 1.4 | 6.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.3 | 6.7 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 1.3 | 10.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 1.3 | 15.6 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 1.2 | 3.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.2 | 9.4 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 1.2 | 9.4 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 1.2 | 9.4 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 1.1 | 3.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 1.1 | 5.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 1.1 | 11.8 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 1.1 | 5.4 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 1.0 | 1.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 1.0 | 9.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.9 | 1.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.9 | 7.0 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.8 | 4.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.8 | 45.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.8 | 86.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.8 | 8.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.7 | 5.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.7 | 6.5 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.7 | 11.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.7 | 2.0 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.6 | 4.8 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.6 | 6.6 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.6 | 2.3 | GO:0042148 | strand invasion(GO:0042148) |
| 0.6 | 4.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.6 | 7.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.6 | 2.9 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.6 | 5.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 2.7 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.5 | 4.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.5 | 3.4 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.4 | 2.6 | GO:0070874 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 | 20.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.4 | 3.7 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.4 | 4.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.4 | 2.0 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.3 | 5.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.3 | 1.3 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.3 | 1.3 | GO:0010459 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.3 | 4.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 2.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.3 | 1.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 | 8.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.3 | 9.1 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.3 | 10.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 5.9 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.3 | 3.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 8.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.3 | 10.1 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 7.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.2 | 16.4 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.2 | 5.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.2 | 4.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.2 | 4.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.2 | 21.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.2 | 14.6 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.2 | 5.0 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 8.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 4.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 3.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 3.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 2.3 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 3.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 3.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 5.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 5.9 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 4.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 4.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 2.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 2.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 7.3 | GO:0042594 | response to starvation(GO:0042594) |
| 0.1 | 1.6 | GO:0051604 | protein maturation(GO:0051604) |
| 0.1 | 4.4 | GO:0097530 | granulocyte migration(GO:0097530) |
| 0.1 | 2.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 3.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 8.0 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.1 | 6.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.3 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 5.3 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 7.6 | GO:0017038 | protein import(GO:0017038) |
| 0.1 | 16.3 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.1 | 26.3 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.1 | 4.4 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 1.6 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 1.9 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 20.4 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 2.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 8.5 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.9 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 3.8 | GO:0030111 | regulation of Wnt signaling pathway(GO:0030111) |
| 0.0 | 24.9 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 1.5 | GO:0006364 | rRNA processing(GO:0006364) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.4 | 37.3 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 2.5 | 10.1 | GO:0031673 | H zone(GO:0031673) |
| 1.5 | 10.5 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 1.5 | 4.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.4 | 5.5 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 1.2 | 14.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.2 | 15.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 1.2 | 16.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.1 | 7.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 1.0 | 14.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.9 | 11.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.8 | 5.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.8 | 17.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.7 | 35.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.6 | 7.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.6 | 71.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.6 | 34.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.5 | 2.0 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.5 | 4.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 2.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.5 | 17.6 | GO:0016605 | PML body(GO:0016605) |
| 0.4 | 4.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 8.1 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.4 | 35.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 15.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.3 | 2.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 24.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.2 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 8.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 6.0 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.2 | 27.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.2 | 23.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 7.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 152.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 11.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 2.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 3.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 5.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 4.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 20.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 80.8 | GO:0005829 | cytosol(GO:0005829) |
| 0.1 | 8.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 1.3 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 6.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 29.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 5.7 | 22.9 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 4.4 | 17.6 | GO:0005521 | lamin binding(GO:0005521) |
| 4.2 | 54.9 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 3.8 | 19.0 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 3.1 | 9.4 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 2.7 | 8.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 2.4 | 14.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 2.3 | 11.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 2.2 | 15.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 2.2 | 6.6 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 2.1 | 8.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 2.1 | 26.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 2.0 | 6.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 1.9 | 40.4 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 1.7 | 12.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.7 | 10.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.5 | 7.6 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 1.4 | 8.6 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 1.4 | 4.1 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.3 | 5.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 1.3 | 30.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 1.2 | 86.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.1 | 3.4 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 1.1 | 7.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 1.1 | 29.6 | GO:0042805 | actinin binding(GO:0042805) |
| 1.1 | 11.8 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.0 | 12.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.0 | 10.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.9 | 6.5 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.9 | 6.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.8 | 3.4 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.8 | 2.5 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.8 | 5.7 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.8 | 8.4 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.8 | 12.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.7 | 51.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.7 | 4.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.7 | 6.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.7 | 2.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.6 | 11.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.6 | 10.8 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.6 | 10.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.6 | 16.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.6 | 110.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.5 | 43.9 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.5 | 4.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 7.0 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.5 | 17.5 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.5 | 2.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 2.6 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.4 | 12.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 1.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.4 | 20.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.4 | 4.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 3.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.4 | 1.5 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.4 | 5.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 10.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 7.0 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.3 | 3.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 4.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.3 | 3.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.3 | 5.1 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.3 | 5.9 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.3 | 2.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 10.1 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 2.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 59.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.2 | 1.6 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 5.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 4.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 5.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 33.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.2 | 35.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 20.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 6.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 8.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 12.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 1.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 3.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 3.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 3.5 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 3.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 6.0 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 2.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 32.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 3.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 2.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 3.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 8.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 3.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 6.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.9 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 3.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 9.5 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 6.0 | GO:0016798 | hydrolase activity, acting on glycosyl bonds(GO:0016798) |
| 0.0 | 19.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 31.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 3.7 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 2.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 16.3 | GO:0038023 | signaling receptor activity(GO:0038023) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 37.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 5.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.4 | 22.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.3 | 10.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 3.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 9.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 8.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 12.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 7.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 21.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 2.2 | 9.0 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 1.7 | 10.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.4 | 75.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 1.4 | 25.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.3 | 15.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 1.3 | 26.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.2 | 8.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.1 | 3.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.9 | 14.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.7 | 53.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.7 | 3.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.7 | 9.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 5.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.5 | 10.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 2.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 2.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 6.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.3 | 5.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 7.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 3.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 6.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.2 | 12.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.2 | 7.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 23.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 4.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 8.6 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |