Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
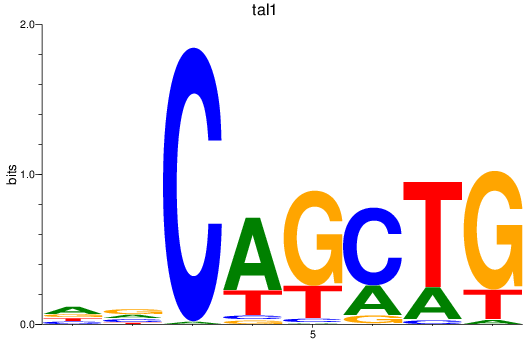
Results for tal1
Z-value: 2.06
Transcription factors associated with tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tal1
|
ENSDARG00000019930 | T-cell acute lymphocytic leukemia 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tal1 | dr11_v1_chr22_+_16535575_16535575 | -0.74 | 2.6e-17 | Click! |
Activity profile of tal1 motif
Sorted Z-values of tal1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61181018 | 35.05 |
ENSDART00000187970
|
pvalb4
|
parvalbumin 4 |
| chr12_+_13256415 | 31.84 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr8_-_1051438 | 26.67 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr7_+_39389273 | 25.74 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr22_+_20720808 | 25.03 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr3_-_61203203 | 23.80 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr18_+_5547185 | 23.53 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr9_+_307863 | 23.10 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr3_-_3209432 | 21.95 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr3_-_1190132 | 21.76 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr18_+_54354 | 19.38 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr11_-_45171139 | 16.93 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr23_+_44614056 | 16.35 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr15_-_551177 | 13.99 |
ENSDART00000066774
ENSDART00000154617 |
tagln
|
transgelin |
| chr3_+_2669813 | 13.85 |
ENSDART00000014205
|
CR388047.1
|
|
| chr2_-_44255537 | 13.69 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr21_-_551014 | 13.55 |
ENSDART00000099252
|
vimr1
|
vimentin-related 1 |
| chr5_-_7829657 | 13.42 |
ENSDART00000158374
|
pdlim5a
|
PDZ and LIM domain 5a |
| chr5_+_338154 | 13.30 |
ENSDART00000191743
|
rnf170
|
ring finger protein 170 |
| chr18_+_5454341 | 13.21 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr7_+_31891110 | 12.84 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr3_+_58833306 | 12.61 |
ENSDART00000113223
|
igl1c3
|
immunoglobulin light 1 constant 3 |
| chr22_-_37738203 | 12.54 |
ENSDART00000143190
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr17_+_53311618 | 11.91 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr20_+_33981946 | 11.86 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr16_+_47308856 | 11.82 |
ENSDART00000170103
|
col28a1b
|
collagen, type XXVIII, alpha 1b |
| chr13_+_51981764 | 11.81 |
ENSDART00000160698
|
klhdc3
|
kelch domain containing 3 |
| chr1_+_50293938 | 11.75 |
ENSDART00000084184
|
aimp1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr1_-_206208 | 11.71 |
ENSDART00000060968
|
adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr24_+_42074143 | 11.59 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr5_+_431994 | 11.36 |
ENSDART00000181692
ENSDART00000170350 |
thap1
|
THAP domain containing, apoptosis associated protein 1 |
| chr14_-_9281232 | 11.31 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr25_-_37501371 | 11.24 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr19_+_48117995 | 11.21 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr23_+_19790962 | 11.16 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr20_-_54377933 | 11.13 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr21_-_217589 | 10.98 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr9_+_33220342 | 10.90 |
ENSDART00000100893
ENSDART00000113451 |
si:ch211-125e6.13
|
si:ch211-125e6.13 |
| chr9_-_105135 | 10.55 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr25_+_3318192 | 10.51 |
ENSDART00000146154
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr13_-_438705 | 10.35 |
ENSDART00000082142
|
CU570800.1
|
|
| chr9_+_32178050 | 10.04 |
ENSDART00000169526
|
coq10b
|
coenzyme Q10B |
| chr17_-_868004 | 10.02 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr4_+_77943184 | 9.94 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr17_+_135590 | 9.65 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr4_-_16354292 | 9.62 |
ENSDART00000139919
|
lum
|
lumican |
| chr9_+_44994214 | 9.55 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr17_-_2584423 | 9.02 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr22_+_18469004 | 8.91 |
ENSDART00000061430
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr6_-_13206255 | 8.89 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr10_+_40633990 | 8.88 |
ENSDART00000190489
ENSDART00000139474 |
si:ch211-238p8.31
|
si:ch211-238p8.31 |
| chr3_+_1209006 | 8.80 |
ENSDART00000158633
|
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr4_-_64703 | 8.64 |
ENSDART00000167851
|
CU856344.1
|
|
| chr7_-_30127082 | 8.59 |
ENSDART00000173749
|
alpk3b
|
alpha-kinase 3b |
| chr1_-_513762 | 8.58 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr17_-_2595736 | 8.57 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_+_53156530 | 8.50 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr17_+_132555 | 8.40 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr5_-_72376973 | 8.32 |
ENSDART00000164969
|
CABZ01100198.1
|
|
| chr17_-_2573021 | 8.29 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_+_53311243 | 8.14 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr25_-_18470695 | 8.00 |
ENSDART00000034377
|
cpa5
|
carboxypeptidase A5 |
| chr23_-_9925568 | 7.89 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr16_+_54588930 | 7.88 |
ENSDART00000159174
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr3_-_5413018 | 7.83 |
ENSDART00000063138
|
vmo1a
|
vitelline membrane outer layer 1 homolog a |
| chr18_-_11729 | 7.75 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr11_-_1392468 | 7.67 |
ENSDART00000004423
|
iars
|
isoleucyl-tRNA synthetase |
| chr18_-_46010 | 7.58 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr24_+_26276805 | 7.56 |
ENSDART00000089749
|
adipoqa
|
adiponectin, C1Q and collagen domain containing, a |
| chr6_+_40629066 | 7.39 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr9_+_33217243 | 7.28 |
ENSDART00000053061
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr13_-_36581875 | 7.26 |
ENSDART00000113204
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr20_+_2460864 | 7.23 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr1_-_52494122 | 7.14 |
ENSDART00000131407
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr9_+_51225345 | 7.08 |
ENSDART00000132896
|
fap
|
fibroblast activation protein, alpha |
| chr11_+_42587900 | 7.07 |
ENSDART00000167529
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr19_-_27588842 | 7.03 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr17_-_2590222 | 6.97 |
ENSDART00000185711
|
CR759892.1
|
|
| chr21_-_45382112 | 6.90 |
ENSDART00000151029
ENSDART00000151335 ENSDART00000151687 ENSDART00000075438 |
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr9_-_23747264 | 6.79 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr24_+_42132962 | 6.75 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr22_-_10541372 | 6.71 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr3_-_1388936 | 6.71 |
ENSDART00000171278
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr1_-_59232267 | 6.69 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr21_-_30293224 | 6.64 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr17_+_20607487 | 6.62 |
ENSDART00000154123
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr21_+_10577527 | 6.59 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr24_-_6029314 | 6.58 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr7_-_6346859 | 6.56 |
ENSDART00000172913
|
si:ch73-368j24.11
|
si:ch73-368j24.11 |
| chr3_-_15154871 | 6.42 |
ENSDART00000147704
|
si:dkey-282h22.5
|
si:dkey-282h22.5 |
| chr1_+_2260407 | 6.28 |
ENSDART00000058876
|
kpnb3
|
karyopherin (importin) beta 3 |
| chr5_+_25762271 | 6.27 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr12_-_656540 | 6.01 |
ENSDART00000172651
|
sult2st2
|
sulfotransferase family 2, cytosolic sulfotransferase 2 |
| chr17_-_2578026 | 5.96 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr3_+_3519607 | 5.94 |
ENSDART00000150922
|
cdc42ep1b
|
CDC42 effector protein (Rho GTPase binding) 1b |
| chr22_-_26353916 | 5.80 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr17_+_584369 | 5.67 |
ENSDART00000165143
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr7_-_6470431 | 5.62 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr14_-_17563773 | 5.60 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor like 1a |
| chr5_+_36895860 | 5.59 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr6_+_59176470 | 5.59 |
ENSDART00000161720
|
shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr17_-_22001303 | 5.55 |
ENSDART00000122190
|
slc22a7b.2
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 2 |
| chr7_-_27033080 | 5.53 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr12_-_48168135 | 5.50 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr3_+_1179601 | 5.50 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr16_+_9495583 | 5.49 |
ENSDART00000150750
ENSDART00000150457 |
pimr208
|
Pim proto-oncogene, serine/threonine kinase, related 208 |
| chr3_+_5504990 | 5.47 |
ENSDART00000143626
|
si:ch73-264i18.2
|
si:ch73-264i18.2 |
| chr19_-_25519612 | 5.40 |
ENSDART00000133150
|
C1GALT1 (1 of many)
|
si:dkey-202e17.1 |
| chr15_+_47525073 | 5.37 |
ENSDART00000067583
|
sidt2
|
SID1 transmembrane family, member 2 |
| chr20_+_2281933 | 5.36 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr11_+_27364338 | 5.32 |
ENSDART00000186759
|
fbln2
|
fibulin 2 |
| chr20_-_49729446 | 5.29 |
ENSDART00000111089
|
filip1b
|
filamin A interacting protein 1b |
| chr3_+_3545825 | 5.27 |
ENSDART00000109060
|
CR589947.1
|
|
| chr3_+_726000 | 5.26 |
ENSDART00000158510
|
dicp1.17
|
diverse immunoglobulin domain-containing protein 1.17 |
| chr3_+_54581987 | 5.15 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr17_+_53439606 | 5.03 |
ENSDART00000154946
|
si:zfos-1714f5.3
|
si:zfos-1714f5.3 |
| chr4_+_5249494 | 4.98 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr11_+_37909654 | 4.96 |
ENSDART00000172211
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr13_+_669177 | 4.96 |
ENSDART00000190085
|
CU462915.1
|
|
| chr25_-_36369057 | 4.84 |
ENSDART00000064400
|
si:ch211-113a14.24
|
si:ch211-113a14.24 |
| chr17_-_681142 | 4.83 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr11_-_44647286 | 4.83 |
ENSDART00000169329
ENSDART00000158939 |
tomm20b
|
translocase of outer mitochondrial membrane 20b |
| chr3_-_41715690 | 4.83 |
ENSDART00000184703
|
BX324110.1
|
|
| chr7_+_1467863 | 4.73 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr25_+_37290206 | 4.71 |
ENSDART00000086474
|
si:dkey-234i14.12
|
si:dkey-234i14.12 |
| chr25_+_15933411 | 4.70 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr19_-_3783141 | 4.67 |
ENSDART00000162260
|
btr19
|
bloodthirsty-related gene family, member 19 |
| chr10_+_40629616 | 4.65 |
ENSDART00000147476
|
CR396590.9
|
|
| chr5_-_15692030 | 4.64 |
ENSDART00000099545
|
CR384085.1
|
|
| chr15_-_34051457 | 4.63 |
ENSDART00000189764
|
VWDE
|
si:dkey-30e9.7 |
| chr25_+_37289088 | 4.58 |
ENSDART00000073438
|
si:dkey-234i14.9
|
si:dkey-234i14.9 |
| chr2_-_37888429 | 4.53 |
ENSDART00000183277
|
mbl2
|
mannose binding lectin 2 |
| chr3_-_3939785 | 4.52 |
ENSDART00000049593
|
unm_sa1506
|
un-named sa1506 |
| chr3_-_34098731 | 4.51 |
ENSDART00000150999
|
ighv5-3
|
immunoglobulin heavy variable 5-3 |
| chr3_+_17537352 | 4.51 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr24_+_35827766 | 4.50 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr19_-_81851 | 4.49 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr7_-_6415991 | 4.45 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr2_+_19633493 | 4.43 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr3_-_34074806 | 4.43 |
ENSDART00000151415
|
ighv8-1
|
immunoglobulin heavy variable 8-1 |
| chr10_-_40536963 | 4.42 |
ENSDART00000184052
ENSDART00000132523 |
taar18e
|
trace amine associated receptor 18e |
| chr3_+_3598555 | 4.41 |
ENSDART00000191152
|
CR589947.3
|
|
| chr11_+_27364059 | 4.37 |
ENSDART00000172883
|
fbln2
|
fibulin 2 |
| chr7_+_5964296 | 4.36 |
ENSDART00000173380
|
si:dkey-23a13.17
|
si:dkey-23a13.17 |
| chr10_-_8294965 | 4.33 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr12_+_33433882 | 4.32 |
ENSDART00000185053
|
fasn
|
fatty acid synthase |
| chr5_+_37785152 | 4.28 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr12_-_48960308 | 4.26 |
ENSDART00000176247
|
CABZ01112647.1
|
|
| chr23_-_44848961 | 4.24 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr1_-_53880639 | 4.19 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr1_-_53468160 | 4.16 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr15_+_28368823 | 4.16 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr1_+_57235896 | 4.12 |
ENSDART00000152621
|
si:dkey-27j5.7
|
si:dkey-27j5.7 |
| chr24_+_5789582 | 4.09 |
ENSDART00000141504
|
BX470065.1
|
|
| chr25_-_35153985 | 4.06 |
ENSDART00000154851
|
zgc:153405
|
zgc:153405 |
| chr1_-_55263736 | 4.06 |
ENSDART00000152504
ENSDART00000152687 |
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr15_-_7598294 | 4.04 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr1_-_59141715 | 4.03 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr8_-_1219815 | 4.03 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr3_-_10970502 | 3.98 |
ENSDART00000127500
|
CR382337.1
|
|
| chr3_-_45848043 | 3.93 |
ENSDART00000055132
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr10_+_40737540 | 3.93 |
ENSDART00000125577
|
taar19a
|
trace amine associated receptor 19a |
| chr15_-_4485828 | 3.91 |
ENSDART00000062868
|
tfdp2
|
transcription factor Dp-2 |
| chr7_+_33132074 | 3.90 |
ENSDART00000073554
|
zgc:153219
|
zgc:153219 |
| chr8_-_27687095 | 3.90 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr3_-_2613990 | 3.89 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr23_-_40895168 | 3.87 |
ENSDART00000061037
|
si:dkey-194e6.1
|
si:dkey-194e6.1 |
| chr4_+_2267641 | 3.85 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr3_-_32590164 | 3.85 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr10_+_26896142 | 3.83 |
ENSDART00000188225
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr7_+_6317866 | 3.82 |
ENSDART00000173397
|
si:ch211-220f21.3
|
si:ch211-220f21.3 |
| chr1_-_55118745 | 3.81 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr25_+_1732838 | 3.80 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr21_+_5531138 | 3.78 |
ENSDART00000163825
|
ly6m6
|
lymphocyte antigen 6 family member M6 |
| chr16_+_32029090 | 3.76 |
ENSDART00000041054
|
tmc4
|
transmembrane channel-like 4 |
| chr25_-_36263115 | 3.75 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr13_-_133520 | 3.74 |
ENSDART00000180874
|
FO904997.1
|
|
| chr25_+_37340722 | 3.72 |
ENSDART00000137025
|
pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr5_+_7564644 | 3.71 |
ENSDART00000192173
|
CABZ01039096.1
|
|
| chr24_-_26310854 | 3.71 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr7_+_35229805 | 3.68 |
ENSDART00000173911
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr14_+_9432627 | 3.68 |
ENSDART00000042727
|
si:dkeyp-86f7.4
|
si:dkeyp-86f7.4 |
| chr23_+_1349277 | 3.68 |
ENSDART00000173133
ENSDART00000179877 |
utrn
|
utrophin |
| chr6_-_35439406 | 3.68 |
ENSDART00000073784
|
rgs5a
|
regulator of G protein signaling 5a |
| chr3_-_61592417 | 3.67 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr5_+_28857969 | 3.66 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr12_-_49166761 | 3.63 |
ENSDART00000189407
|
acadsb
|
acyl-CoA dehydrogenase short/branched chain |
| chr10_+_406146 | 3.63 |
ENSDART00000145124
|
dact3a
|
dishevelled-binding antagonist of beta-catenin 3a |
| chr3_-_34037064 | 3.63 |
ENSDART00000151716
|
ighv9-4
|
immunoglobulin heavy variable 9-4 |
| chr3_-_2623176 | 3.62 |
ENSDART00000179792
ENSDART00000123512 |
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr1_+_55703120 | 3.61 |
ENSDART00000141089
|
adgre6
|
adhesion G protein-coupled receptor E6 |
| chr7_-_8470860 | 3.58 |
ENSDART00000172793
|
loc564481
|
hypothetical protein LOC564481 |
| chr22_+_5971404 | 3.55 |
ENSDART00000122153
|
BX322587.1
|
|
| chr1_+_277731 | 3.54 |
ENSDART00000133431
|
cenpe
|
centromere protein E |
| chr25_-_36261836 | 3.53 |
ENSDART00000179411
|
dus2
|
dihydrouridine synthase 2 |
| chr5_+_41322783 | 3.53 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr14_+_17382803 | 3.53 |
ENSDART00000040383
|
si:ch211-255i20.3
|
si:ch211-255i20.3 |
| chr4_+_8638622 | 3.53 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr13_-_301309 | 3.50 |
ENSDART00000131747
|
chs1
|
chitin synthase 1 |
| chr22_+_8272393 | 3.46 |
ENSDART00000121602
|
CABZ01077220.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of tal1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 4.6 | 23.1 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 4.6 | 13.7 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 4.0 | 11.9 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 3.9 | 11.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 2.6 | 12.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 2.5 | 7.6 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 2.4 | 26.7 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 2.3 | 18.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 1.9 | 31.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 1.9 | 5.6 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 1.8 | 8.9 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 1.8 | 31.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 1.5 | 7.3 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 1.4 | 8.5 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 1.0 | 9.9 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 1.0 | 4.8 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.9 | 17.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.9 | 6.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) embryonic cleavage(GO:0040016) |
| 0.9 | 6.6 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.9 | 2.8 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.8 | 2.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.8 | 4.0 | GO:1990592 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.8 | 3.9 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.8 | 4.5 | GO:0042745 | temperature homeostasis(GO:0001659) circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) regulation of transmission of nerve impulse(GO:0051969) |
| 0.7 | 20.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.7 | 3.5 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.7 | 11.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.7 | 3.5 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.7 | 5.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.7 | 11.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.7 | 2.0 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.7 | 3.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.7 | 11.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.6 | 2.4 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.6 | 25.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.6 | 5.9 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.6 | 2.3 | GO:0042148 | strand invasion(GO:0042148) |
| 0.6 | 1.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.6 | 5.5 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.5 | 2.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.5 | 28.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.5 | 7.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.5 | 10.0 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 7.5 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.5 | 2.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.5 | 5.6 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.5 | 4.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 10.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.4 | 3.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 2.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 2.4 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.4 | 2.7 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 8.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 6.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 2.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 | 10.1 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.3 | 2.7 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.3 | 2.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) regulation of stem cell proliferation(GO:0072091) |
| 0.3 | 2.3 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.3 | 4.4 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.3 | 1.5 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.3 | 1.8 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.3 | 1.5 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.3 | 3.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 2.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 5.3 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.3 | 0.5 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.3 | 0.8 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.3 | 3.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 20.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.2 | 9.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 1.0 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 1.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 1.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 2.3 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.2 | 6.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 2.9 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.2 | 2.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 | 5.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 3.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 1.6 | GO:0007197 | regulation of smooth muscle contraction(GO:0006940) adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 0.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 0.7 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.2 | 3.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 1.0 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 6.0 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 3.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 4.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 8.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 3.1 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 4.7 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 3.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 1.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 3.7 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 0.4 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 2.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 8.2 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 4.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 6.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 27.2 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 3.8 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.1 | 4.0 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.9 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.8 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 1.2 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 25.3 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.1 | 0.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 9.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.7 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 3.3 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.1 | 0.7 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 1.1 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.7 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 4.3 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.1 | 1.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.3 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 2.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 4.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 1.0 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 1.9 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 2.7 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 1.7 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 6.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.1 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.2 | GO:0032941 | body fluid secretion(GO:0007589) secretion by tissue(GO:0032941) mucus secretion(GO:0070254) |
| 0.0 | 0.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.7 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.5 | GO:0014020 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) |
| 0.0 | 0.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.1 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.7 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 3.4 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 2.1 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.4 | 23.1 | GO:0030315 | T-tubule(GO:0030315) |
| 1.3 | 17.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.3 | 31.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 1.3 | 8.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.3 | 26.7 | GO:0031430 | M band(GO:0031430) |
| 1.2 | 7.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.1 | 8.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 1.1 | 4.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.9 | 7.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.7 | 5.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.6 | 25.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 2.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.5 | 16.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.5 | 28.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.5 | 3.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.5 | 2.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 4.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.4 | 3.5 | GO:0030428 | cell septum(GO:0030428) |
| 0.4 | 17.3 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.4 | 4.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.4 | 17.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.3 | 2.4 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.3 | 28.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.3 | 26.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 2.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 5.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.2 | 3.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.2 | 5.6 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 1.5 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 6.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 3.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 4.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.9 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 35.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 6.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.7 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 2.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 5.3 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 34.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 10.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 7.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 2.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 2.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 4.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.3 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.5 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 3.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 4.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.6 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.1 | 11.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.1 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 25.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 23.5 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 3.9 | 11.7 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 3.0 | 11.9 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 2.9 | 11.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 2.9 | 31.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 2.5 | 7.4 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 2.3 | 18.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 2.2 | 8.9 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 1.9 | 31.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.9 | 5.6 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 1.5 | 7.3 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 1.4 | 13.7 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 1.3 | 3.9 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 1.3 | 19.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.2 | 7.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 1.2 | 12.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.1 | 25.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 1.1 | 10.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.1 | 3.3 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 1.0 | 3.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 1.0 | 4.0 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.8 | 3.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.8 | 18.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.8 | 2.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.7 | 10.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.7 | 2.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.7 | 2.0 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.7 | 13.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.7 | 11.1 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.6 | 7.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.6 | 3.8 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.6 | 10.2 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.6 | 1.8 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.6 | 1.7 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.5 | 28.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.5 | 3.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.5 | 3.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.5 | 6.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 3.5 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.4 | 5.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 1.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.4 | 1.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.4 | 27.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.4 | 7.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.4 | 11.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.4 | 6.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.4 | 1.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 2.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.3 | 9.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 10.2 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.3 | 2.9 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.3 | 1.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 2.7 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 6.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.3 | 4.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.3 | 1.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.3 | 1.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.3 | 1.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 3.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 5.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 2.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 7.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.2 | 0.9 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 6.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.7 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 8.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 2.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 5.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 7.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 5.8 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 3.6 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.2 | 3.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 13.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 1.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 2.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.7 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 2.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 16.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.3 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 3.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 3.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 3.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.3 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 8.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.6 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.5 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 6.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 1.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 4.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 2.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 11.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 48.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 20.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 28.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 5.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.5 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 2.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 6.7 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 6.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 8.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 3.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 6.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 14.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 6.0 | GO:0008289 | lipid binding(GO:0008289) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.4 | 11.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 10.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.3 | 1.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 9.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 32.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 9.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 8.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 11.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 4.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 3.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.8 | 16.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.8 | 21.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.5 | 3.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.5 | 3.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.5 | 7.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 4.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 4.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 4.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 11.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 1.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 2.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 2.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.2 | 3.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 4.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 6.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 5.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.8 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 3.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 5.0 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 1.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 3.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 3.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 4.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |