Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
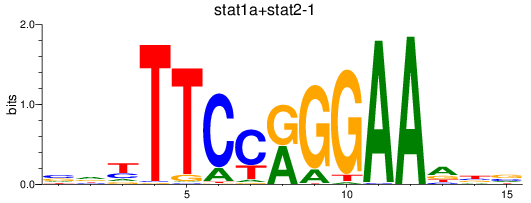
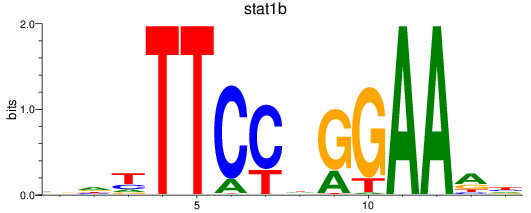
Results for stat1a+stat2-1_stat1b
Z-value: 1.55
Transcription factors associated with stat1a+stat2-1_stat1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat1a
|
ENSDARG00000006266 | signal transducer and activator of transcription 1a |
|
stat2-1
|
ENSDARG00000031647 | signal transducer and activator of transcription 2 |
|
stat1b
|
ENSDARG00000076182 | signal transducer and activator of transcription 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat1b | dr11_v1_chr9_+_41224100_41224100 | 0.59 | 3.0e-10 | Click! |
| stat2 | dr11_v1_chr6_-_39158953_39158953 | 0.31 | 2.4e-03 | Click! |
| stat1a | dr11_v1_chr22_+_12798569_12798569 | 0.06 | 5.9e-01 | Click! |
Activity profile of stat1a+stat2-1_stat1b motif
Sorted Z-values of stat1a+stat2-1_stat1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_36834505 | 23.63 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr21_+_45626136 | 22.46 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr16_+_38394371 | 18.11 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr15_+_20239141 | 17.93 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr14_+_29975268 | 17.91 |
ENSDART00000172985
|
cyp4v7
|
cytochrome P450, family 4, subfamily V, polypeptide 7 |
| chr3_-_44059902 | 16.88 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr6_+_22068589 | 16.73 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr20_+_23440632 | 16.27 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr6_+_41503854 | 16.01 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr17_-_10838434 | 15.86 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr9_-_32813177 | 15.69 |
ENSDART00000012694
|
mxc
|
myxovirus (influenza virus) resistance C |
| chr6_-_8498908 | 15.00 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr6_+_28294113 | 14.40 |
ENSDART00000136898
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr5_-_42878178 | 14.05 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr15_-_29598679 | 12.97 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr23_-_31763753 | 12.90 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr11_-_17964525 | 12.77 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr10_+_17776981 | 12.76 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr16_+_42772678 | 12.72 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr11_+_14286160 | 12.68 |
ENSDART00000166236
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr15_-_32383340 | 12.50 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr6_-_8498676 | 12.50 |
ENSDART00000148627
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr10_-_25823258 | 12.49 |
ENSDART00000064327
|
ftr54
|
finTRIM family, member 54 |
| chr14_+_11457500 | 12.34 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr18_-_14860435 | 12.16 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr2_+_42005217 | 11.78 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr1_-_10071422 | 11.42 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr14_-_30747686 | 11.23 |
ENSDART00000008373
|
fosl1a
|
FOS-like antigen 1a |
| chr14_+_33329420 | 11.09 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr23_+_44611864 | 11.00 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr2_+_26288301 | 10.84 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr9_+_41218967 | 10.76 |
ENSDART00000000280
ENSDART00000145674 |
stat1b
|
signal transducer and activator of transcription 1b |
| chr8_+_47099033 | 10.70 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr21_+_20771082 | 10.69 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr16_-_36099492 | 10.67 |
ENSDART00000180905
|
CU499336.2
|
|
| chr23_-_5101847 | 10.09 |
ENSDART00000122240
|
etv7
|
ets variant 7 |
| chr11_-_8167799 | 9.66 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr6_+_21202639 | 9.51 |
ENSDART00000083126
|
cidec
|
cell death-inducing DFFA-like effector c |
| chr21_-_27338639 | 9.41 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr11_+_13224281 | 9.15 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr16_-_31675669 | 8.94 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr24_-_38113208 | 8.89 |
ENSDART00000079003
|
crp
|
c-reactive protein, pentraxin-related |
| chr12_-_46228023 | 8.83 |
ENSDART00000153455
|
si:ch211-226h7.6
|
si:ch211-226h7.6 |
| chr19_-_5699703 | 8.82 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr20_-_25551676 | 8.78 |
ENSDART00000063081
|
cyp2ad3
|
cytochrome P450, family 2, subfamily AD, polypeptide 3 |
| chr1_-_9195629 | 8.62 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr14_-_32631519 | 8.49 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase phospholipid transporting 11C |
| chr16_-_27749172 | 8.48 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr1_-_49225890 | 8.42 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr14_-_21123551 | 8.26 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr15_-_3282220 | 8.25 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr11_-_22303678 | 8.25 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr2_-_10098191 | 8.18 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr3_+_36646054 | 8.16 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr14_+_16345003 | 8.05 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr25_-_17395315 | 7.99 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr5_-_57624425 | 7.99 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr7_+_73295890 | 7.74 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr18_+_30567945 | 7.73 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr19_+_17385561 | 7.70 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr15_+_6661343 | 7.70 |
ENSDART00000160136
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr6_+_39923052 | 7.58 |
ENSDART00000149019
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr22_+_22004082 | 7.52 |
ENSDART00000148375
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr11_-_287670 | 7.49 |
ENSDART00000035737
|
slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr2_+_42005475 | 7.46 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr15_-_29598444 | 7.46 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr7_-_28658143 | 7.45 |
ENSDART00000173556
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr23_-_5783421 | 7.43 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr8_-_38406831 | 7.36 |
ENSDART00000112991
ENSDART00000191445 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr16_+_40954481 | 7.32 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr7_+_26029672 | 7.32 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr5_-_42071505 | 7.30 |
ENSDART00000137224
ENSDART00000193721 |
cxcl11.7
cenpv
|
chemokine (C-X-C motif) ligand 11, duplicate 7 centromere protein V |
| chr15_-_32383529 | 7.23 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr1_-_8020589 | 7.19 |
ENSDART00000143881
|
si:dkeyp-9d4.2
|
si:dkeyp-9d4.2 |
| chr3_+_26813058 | 7.13 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr10_+_13209580 | 7.00 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr7_+_12950507 | 7.00 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr20_-_37813863 | 6.96 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr9_+_40825065 | 6.84 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr5_-_42883761 | 6.77 |
ENSDART00000167374
|
BX323596.2
|
|
| chr19_-_24757231 | 6.69 |
ENSDART00000128177
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr24_-_38197040 | 6.68 |
ENSDART00000137949
ENSDART00000105639 |
igic1s1
|
immunoglobulin light iota constant 1, s1 |
| chr20_+_52458765 | 6.57 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr5_+_57658898 | 6.53 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr16_+_40560622 | 6.53 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr14_+_33329761 | 6.51 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr9_-_12443726 | 6.43 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr5_-_25583125 | 6.24 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr2_-_42035250 | 6.20 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr7_-_28696556 | 6.19 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr15_+_37412883 | 6.16 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr5_+_1911814 | 6.10 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr3_-_24337814 | 6.02 |
ENSDART00000153553
|
csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr23_-_33558161 | 6.01 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr3_-_49110710 | 5.97 |
ENSDART00000160404
|
trim35-12
|
tripartite motif containing 35-12 |
| chr8_+_19548985 | 5.88 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr13_+_30696286 | 5.87 |
ENSDART00000192411
|
cxcl18a.1
|
chemokine (C-X-C motif) ligand 18a, duplicate 1 |
| chr8_+_8936912 | 5.84 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr5_-_30620625 | 5.78 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr14_-_36799280 | 5.76 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr2_+_4402765 | 5.69 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr2_+_4207209 | 5.64 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr13_+_13681681 | 5.57 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr21_-_22676323 | 5.44 |
ENSDART00000167392
|
gig2h
|
grass carp reovirus (GCRV)-induced gene 2h |
| chr7_+_39706004 | 5.40 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr3_-_24338163 | 5.39 |
ENSDART00000115283
|
csf2rb
|
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr5_-_25582721 | 5.39 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr17_+_16046132 | 5.35 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr9_-_23922011 | 5.34 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr20_-_3238110 | 5.26 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr20_+_25552057 | 5.24 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr9_-_23922778 | 5.20 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr18_-_16801033 | 5.20 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr11_-_27962757 | 5.16 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr3_-_32275975 | 5.13 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr23_+_10469955 | 5.11 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr18_-_40773413 | 5.10 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr16_+_54513267 | 5.04 |
ENSDART00000161923
|
CABZ01088508.1
|
|
| chr12_-_23365737 | 4.95 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr1_-_6085750 | 4.92 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr23_+_1216215 | 4.91 |
ENSDART00000165957
|
utrn
|
utrophin |
| chr17_+_16046314 | 4.89 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr13_-_37631092 | 4.89 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr21_-_10886709 | 4.83 |
ENSDART00000134408
|
si:dkey-277m11.2
|
si:dkey-277m11.2 |
| chr22_-_16180467 | 4.72 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr8_-_31701157 | 4.72 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr24_-_19719240 | 4.69 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr11_+_13223625 | 4.65 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr7_+_41313568 | 4.62 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr10_+_24692076 | 4.60 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr7_-_20241346 | 4.50 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr6_+_102506 | 4.46 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr13_-_40238813 | 4.46 |
ENSDART00000044963
|
loxl4
|
lysyl oxidase-like 4 |
| chr24_-_25256670 | 4.44 |
ENSDART00000126270
|
hhla2b.1
|
HERV-H LTR-associating 2b, tandem duplicate 1 |
| chr2_+_22659787 | 4.44 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr5_+_29820266 | 4.41 |
ENSDART00000146331
ENSDART00000098315 |
f11r.2
|
F11 receptor, tandem duplicate 2 |
| chr7_+_26545502 | 4.39 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr4_+_76735113 | 4.39 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr9_-_43644261 | 4.39 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr11_+_16040517 | 4.37 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr10_-_41352502 | 4.34 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr3_+_32571929 | 4.31 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr18_+_13315739 | 4.30 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr7_+_26167420 | 4.19 |
ENSDART00000173941
|
si:ch211-196f2.6
|
si:ch211-196f2.6 |
| chr20_+_36812368 | 4.13 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr19_+_43359075 | 4.10 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr19_-_35596207 | 4.09 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr22_+_7480465 | 4.03 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr17_+_24821627 | 4.01 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr17_+_24064014 | 4.01 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr9_-_54304684 | 3.99 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr19_+_48176745 | 3.98 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr21_-_34844059 | 3.96 |
ENSDART00000136402
|
zgc:56585
|
zgc:56585 |
| chr23_+_33413804 | 3.96 |
ENSDART00000142328
|
rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr21_-_20932603 | 3.95 |
ENSDART00000138155
ENSDART00000079709 |
c6
|
complement component 6 |
| chr17_-_2834764 | 3.86 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr11_-_11882982 | 3.84 |
ENSDART00000190853
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr3_+_22381955 | 3.82 |
ENSDART00000185315
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr17_-_12498096 | 3.81 |
ENSDART00000149551
ENSDART00000105215 ENSDART00000191207 |
emilin1b
|
elastin microfibril interfacer 1b |
| chr25_-_21822426 | 3.79 |
ENSDART00000151993
|
zgc:158222
|
zgc:158222 |
| chr11_+_30321116 | 3.79 |
ENSDART00000187921
ENSDART00000127075 |
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr6_+_19948043 | 3.79 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr21_-_34844316 | 3.78 |
ENSDART00000029708
|
zgc:56585
|
zgc:56585 |
| chr14_+_30774894 | 3.66 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr17_+_6563307 | 3.65 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr4_-_18436899 | 3.64 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr5_+_25317061 | 3.56 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr17_+_10318071 | 3.53 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr12_-_22379421 | 3.48 |
ENSDART00000187875
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr24_-_11905911 | 3.47 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr6_+_18321627 | 3.45 |
ENSDART00000183107
ENSDART00000189715 |
card14
|
caspase recruitment domain family, member 14 |
| chr9_-_9282519 | 3.42 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr7_+_32369463 | 3.38 |
ENSDART00000180544
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr12_-_3133483 | 3.37 |
ENSDART00000015092
|
col1a1b
|
collagen, type I, alpha 1b |
| chr5_+_29160324 | 3.34 |
ENSDART00000137324
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr19_+_43780970 | 3.34 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr10_+_20608676 | 3.27 |
ENSDART00000140141
|
si:dkey-81j8.6
|
si:dkey-81j8.6 |
| chr7_+_41314862 | 3.25 |
ENSDART00000185198
|
zgc:165532
|
zgc:165532 |
| chr16_-_45398408 | 3.24 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr7_+_32369026 | 3.23 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr10_-_11385155 | 3.21 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr21_-_22715297 | 3.20 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr10_-_4375190 | 3.19 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr21_-_20840714 | 3.17 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr6_+_13046720 | 3.17 |
ENSDART00000165896
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr21_-_8085635 | 3.13 |
ENSDART00000082790
|
si:dkey-163m14.2
|
si:dkey-163m14.2 |
| chr20_-_23440955 | 3.09 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr14_-_52433292 | 3.09 |
ENSDART00000164272
|
rest
|
RE1-silencing transcription factor |
| chr22_+_20427170 | 3.08 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr3_-_57779801 | 3.07 |
ENSDART00000074285
|
syngr2a
|
synaptogyrin 2a |
| chr20_+_21595244 | 3.06 |
ENSDART00000010643
|
esr2a
|
estrogen receptor 2a |
| chr22_-_13165186 | 3.06 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr20_-_33704753 | 3.05 |
ENSDART00000157427
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr12_-_16990896 | 3.04 |
ENSDART00000152402
|
ifit12
|
interferon-induced protein with tetratricopeptide repeats 12 |
| chr12_-_46252062 | 3.03 |
ENSDART00000153223
|
si:ch211-226h7.5
|
si:ch211-226h7.5 |
| chr13_-_36798204 | 3.01 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr8_-_38159805 | 3.00 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr4_+_73215536 | 2.94 |
ENSDART00000174290
|
LO018260.2
|
Danio rerio protein NLRC3-like (LOC101883187), mRNA. |
| chr24_-_5932982 | 2.93 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr6_+_37655078 | 2.88 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat1a+stat2-1_stat1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 27.5 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 6.0 | 18.1 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 6.0 | 17.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 5.6 | 16.7 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 4.3 | 12.9 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 4.2 | 16.9 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 4.0 | 27.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 3.4 | 13.8 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 3.2 | 9.7 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 3.2 | 15.9 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 2.8 | 8.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 2.7 | 10.8 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 2.6 | 7.7 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 2.4 | 11.9 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 2.3 | 13.6 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 2.1 | 10.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 2.1 | 8.5 | GO:0015677 | copper ion import(GO:0015677) |
| 2.1 | 16.6 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 1.9 | 11.6 | GO:0045064 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) T-helper 2 cell differentiation(GO:0045064) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.9 | 9.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 1.9 | 22.5 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 1.9 | 7.5 | GO:0010039 | response to iron ion(GO:0010039) |
| 1.9 | 5.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.7 | 8.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 1.7 | 10.2 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 1.4 | 4.2 | GO:0002831 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) regulation of response to biotic stimulus(GO:0002831) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.4 | 11.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 1.3 | 7.9 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 1.1 | 6.6 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 1.0 | 3.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) negative regulation of cell maturation(GO:1903430) |
| 1.0 | 6.0 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 1.0 | 3.0 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 1.0 | 10.8 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.8 | 2.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.8 | 11.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.7 | 2.2 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.7 | 2.9 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.7 | 2.1 | GO:0045226 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.7 | 2.8 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.7 | 2.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.6 | 1.9 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.6 | 1.9 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.6 | 7.3 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.5 | 14.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.5 | 3.5 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.5 | 2.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.5 | 16.9 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.5 | 5.8 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.5 | 1.8 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.4 | 3.1 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.4 | 8.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.4 | 17.2 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.4 | 5.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.4 | 21.3 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.4 | 9.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.4 | 5.8 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.4 | 3.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.3 | 5.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.3 | 4.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.3 | 4.1 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.3 | 7.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 30.4 | GO:0072376 | protein activation cascade(GO:0072376) |
| 0.3 | 2.2 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 3.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 4.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 3.1 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.3 | 6.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.3 | 2.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 0.8 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.3 | 1.3 | GO:2000515 | negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.3 | 17.6 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.2 | 13.4 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.2 | 0.7 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.2 | 5.7 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.2 | 2.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 7.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 2.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.2 | 13.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 2.0 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 | 8.1 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.2 | 10.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 6.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 4.3 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.2 | 1.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 2.4 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.2 | 0.5 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 6.2 | GO:0051896 | regulation of protein kinase B signaling(GO:0051896) |
| 0.1 | 3.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) regulation of cell junction assembly(GO:1901888) |
| 0.1 | 1.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 3.4 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 4.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 7.1 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.6 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 2.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 4.3 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 1.3 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 1.4 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.1 | 4.7 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 7.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 2.0 | GO:0002685 | regulation of leukocyte migration(GO:0002685) |
| 0.1 | 4.2 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 10.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 6.5 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 0.8 | GO:0055062 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 8.2 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.1 | 4.0 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.1 | 3.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.9 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 21.0 | GO:0009116 | nucleoside metabolic process(GO:0009116) |
| 0.1 | 1.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 1.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 2.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.2 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 4.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 2.1 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.1 | 1.0 | GO:0009988 | cell-cell recognition(GO:0009988) sperm-egg recognition(GO:0035036) |
| 0.1 | 0.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 4.0 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 2.7 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 10.4 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 4.4 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 2.1 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.9 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 3.8 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.4 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 19.9 | GO:0042981 | regulation of apoptotic process(GO:0042981) |
| 0.0 | 2.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 2.5 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 4.7 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 1.4 | GO:0021536 | diencephalon development(GO:0021536) |
| 0.0 | 0.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 4.1 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.9 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.3 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 2.9 | 8.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 2.0 | 15.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.9 | 9.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 1.4 | 11.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.8 | 15.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.8 | 11.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.8 | 44.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.8 | 3.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.7 | 4.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.6 | 2.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.5 | 7.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 24.1 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.3 | 2.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 0.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 3.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 11.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 2.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 2.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 23.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 11.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 15.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 7.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 7.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 5.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 3.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.6 | GO:0030669 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 1.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 7.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 9.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 7.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 8.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 5.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 16.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 55.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 9.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 3.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 6.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.4 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 2.2 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.0 | 1.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 4.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 27.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 5.6 | 16.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 3.4 | 13.8 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 3.2 | 15.9 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 2.6 | 23.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 2.5 | 7.5 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 2.1 | 6.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 2.1 | 10.7 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 2.1 | 8.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 2.1 | 8.4 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.9 | 11.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.5 | 7.7 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.4 | 11.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.3 | 44.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 1.2 | 9.7 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 1.0 | 9.6 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.9 | 8.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.9 | 2.7 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.8 | 4.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.7 | 2.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.7 | 2.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.7 | 2.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.6 | 7.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 6.9 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.6 | 6.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.5 | 2.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.5 | 3.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 5.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 10.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 54.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 1.9 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.5 | 6.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 3.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.4 | 5.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 2.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.4 | 3.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.4 | 7.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 17.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 3.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.4 | 1.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 6.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.4 | 2.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 3.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.3 | 1.0 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.3 | 12.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 2.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 21.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.3 | 5.9 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.3 | 2.4 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 12.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.3 | 5.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.3 | 4.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 7.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 0.7 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.2 | 1.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 5.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 3.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 27.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.8 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 2.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 3.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 17.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.2 | 2.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 2.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 1.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 2.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 8.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 7.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 18.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 8.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 2.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 4.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 6.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.6 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 8.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 4.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 8.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 4.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 3.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.8 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 13.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.8 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 3.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 2.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.3 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 0.2 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 6.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 16.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 73.9 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 5.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 5.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 12.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 9.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 16.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.8 | 11.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.7 | 11.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.6 | 7.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.6 | 4.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.5 | 4.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.5 | 6.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.5 | 3.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 17.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.4 | 7.7 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.4 | 14.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 7.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 7.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.3 | 3.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 7.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 3.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 5.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 9.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 4.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 17.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 1.2 | 19.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 1.1 | 33.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 1.0 | 5.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.0 | 11.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.8 | 6.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.6 | 1.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.5 | 7.1 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.5 | 11.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 11.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.4 | 4.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 3.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 7.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 15.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 10.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 3.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 2.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 2.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 7.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 6.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 4.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.6 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 11.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 3.8 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |