Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
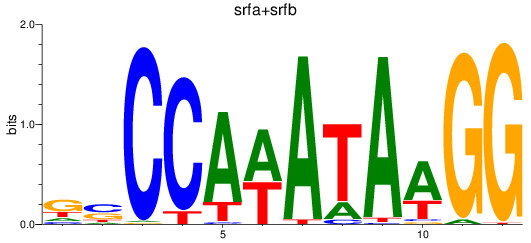
Results for srfa+srfb
Z-value: 2.99
Transcription factors associated with srfa+srfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
srfa
|
ENSDARG00000053918 | serum response factor a |
|
srfb
|
ENSDARG00000102867 | serum response factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| srfb | dr11_v1_chr13_-_51922290_51922290 | 0.64 | 6.3e-12 | Click! |
| srfa | dr11_v1_chr22_+_35089031_35089031 | -0.30 | 3.1e-03 | Click! |
Activity profile of srfa+srfb motif
Sorted Z-values of srfa+srfb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_9980318 | 81.33 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr12_-_26064480 | 79.44 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr12_-_26064105 | 71.08 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr13_+_24279021 | 64.83 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr1_+_53945934 | 61.72 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr15_-_551177 | 54.42 |
ENSDART00000066774
ENSDART00000154617 |
tagln
|
transgelin |
| chr2_+_30916188 | 45.09 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr22_-_15602760 | 44.63 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr23_+_42813415 | 40.06 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr23_+_44611864 | 37.42 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr7_-_71758613 | 35.57 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr10_+_6318227 | 34.87 |
ENSDART00000170872
ENSDART00000162428 ENSDART00000158994 |
tpm2
|
tropomyosin 2 (beta) |
| chr8_-_11229523 | 33.81 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr17_-_25326296 | 32.46 |
ENSDART00000168822
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr13_+_33606739 | 32.14 |
ENSDART00000026464
|
cfl1l
|
cofilin 1 (non-muscle), like |
| chr14_+_49251331 | 31.88 |
ENSDART00000148882
|
anxa6
|
annexin A6 |
| chr12_-_20373058 | 31.25 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr8_+_40628926 | 31.19 |
ENSDART00000163598
|
dusp2
|
dual specificity phosphatase 2 |
| chr22_-_15602593 | 30.88 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr20_+_40150612 | 30.82 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr20_+_53577502 | 30.09 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr21_+_25765734 | 29.95 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr2_+_15100742 | 28.80 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr5_+_37087583 | 28.15 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr7_+_29952169 | 27.49 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr2_-_24269911 | 27.29 |
ENSDART00000099532
|
myh7
|
myosin heavy chain 7 |
| chr19_+_46158078 | 26.67 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr20_-_29420713 | 26.50 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr22_+_336256 | 24.68 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr6_+_1787160 | 24.29 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr5_-_32292965 | 24.03 |
ENSDART00000183522
ENSDART00000131983 |
myhz1.2
|
myosin, heavy polypeptide 1.2, skeletal muscle |
| chr17_-_15611744 | 23.83 |
ENSDART00000010496
|
fhl5
|
four and a half LIM domains 5 |
| chr5_-_32309129 | 23.48 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr19_+_348729 | 23.17 |
ENSDART00000114284
|
mcl1a
|
MCL1, BCL2 family apoptosis regulator a |
| chr1_+_9994811 | 23.02 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr14_+_34514336 | 22.01 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr1_-_14332283 | 20.57 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr13_+_29238850 | 19.46 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr13_+_8840772 | 19.40 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr7_-_71758307 | 19.11 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr12_+_17100021 | 18.39 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr9_+_6587056 | 17.80 |
ENSDART00000193421
|
fhl2a
|
four and a half LIM domains 2a |
| chr5_+_37785152 | 17.75 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr17_-_20118145 | 17.62 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr21_-_17603182 | 17.40 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr10_-_373575 | 17.19 |
ENSDART00000114487
|
DMPK
|
DM1 protein kinase |
| chr25_+_33192796 | 17.09 |
ENSDART00000125892
ENSDART00000121680 ENSDART00000014851 |
TPM1 (1 of many)
|
zgc:171719 |
| chr1_+_1805294 | 17.07 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr25_+_33192404 | 16.87 |
ENSDART00000193592
|
TPM1 (1 of many)
|
zgc:171719 |
| chr9_+_6587364 | 16.67 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr2_-_24270062 | 16.47 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr14_-_32824380 | 15.62 |
ENSDART00000172791
ENSDART00000105745 |
inppl1b
|
inositol polyphosphate phosphatase-like 1b |
| chr10_+_15603082 | 15.31 |
ENSDART00000024450
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr18_+_36770166 | 14.64 |
ENSDART00000078151
|
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr1_-_8651718 | 14.47 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr1_-_8652648 | 14.07 |
ENSDART00000138324
ENSDART00000141407 ENSDART00000054987 |
actb1
|
actin, beta 1 |
| chr3_-_22228602 | 13.85 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr7_+_29951997 | 13.26 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr3_+_58167288 | 13.23 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr14_+_164556 | 12.72 |
ENSDART00000185606
|
WDR1
|
WD repeat domain 1 |
| chr17_-_43666166 | 12.69 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr18_-_41650648 | 12.60 |
ENSDART00000024087
|
fzd9b
|
frizzled class receptor 9b |
| chr6_-_45905746 | 12.15 |
ENSDART00000025428
|
epha2a
|
eph receptor A2 a |
| chr23_-_35064785 | 12.02 |
ENSDART00000172240
|
BX294434.1
|
|
| chr22_-_17458070 | 11.60 |
ENSDART00000139658
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr20_-_14875308 | 11.32 |
ENSDART00000141290
|
dnm3a
|
dynamin 3a |
| chr20_-_46554440 | 11.07 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr1_-_69444 | 10.58 |
ENSDART00000166954
|
si:zfos-1011f11.1
|
si:zfos-1011f11.1 |
| chr18_+_36769758 | 10.38 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr6_+_41191482 | 10.08 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr17_+_25849332 | 10.02 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr13_+_23132666 | 9.90 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr14_-_30747686 | 9.04 |
ENSDART00000008373
|
fosl1a
|
FOS-like antigen 1a |
| chr2_-_24554416 | 8.96 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr18_-_46258612 | 8.71 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr7_-_18168493 | 8.62 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr5_-_41845116 | 8.39 |
ENSDART00000112382
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr1_-_37383741 | 7.92 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr16_-_17347727 | 7.85 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr10_-_7892192 | 7.77 |
ENSDART00000145480
|
smtna
|
smoothelin a |
| chr3_+_1211242 | 7.77 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr3_+_32410746 | 7.69 |
ENSDART00000025496
|
rras
|
RAS related |
| chr20_-_31018569 | 7.60 |
ENSDART00000136039
|
fndc1
|
fibronectin type III domain containing 1 |
| chr3_-_18410968 | 7.43 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr19_+_43715911 | 7.37 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr20_+_1996202 | 7.36 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr13_-_3155243 | 6.66 |
ENSDART00000139183
ENSDART00000050934 |
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr7_-_53117131 | 6.52 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr6_+_41200500 | 6.43 |
ENSDART00000000678
|
opn1mw4
|
opsin 1 (cone pigments), medium-wave-sensitive, 4 |
| chr11_+_30647545 | 6.32 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr20_+_46572550 | 6.16 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr11_+_6115621 | 6.01 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr13_-_9598320 | 5.94 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr3_-_40529782 | 5.64 |
ENSDART00000055194
ENSDART00000141737 |
actb2
|
actin, beta 2 |
| chr21_+_30043054 | 5.63 |
ENSDART00000065448
|
fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr18_+_35407921 | 5.59 |
ENSDART00000057733
|
zgc:153119
|
zgc:153119 |
| chr13_-_40398556 | 5.55 |
ENSDART00000191921
|
nkx3.3
|
NK3 homeobox 3 |
| chr4_-_16644708 | 5.51 |
ENSDART00000042307
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr16_-_31754102 | 5.48 |
ENSDART00000185043
|
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr21_-_31286428 | 5.45 |
ENSDART00000185334
|
ca4c
|
carbonic anhydrase IV c |
| chr7_-_66868543 | 5.42 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr16_+_28383758 | 5.36 |
ENSDART00000059038
ENSDART00000141061 |
itga8
|
integrin, alpha 8 |
| chr9_-_44948488 | 5.33 |
ENSDART00000059228
|
vil1
|
villin 1 |
| chr24_+_31277360 | 5.32 |
ENSDART00000165993
|
f3a
|
coagulation factor IIIa |
| chr19_-_20403845 | 5.23 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr23_+_19655301 | 5.14 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr21_+_8533533 | 5.11 |
ENSDART00000077924
|
FO834888.1
|
|
| chr8_+_26059677 | 5.10 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr14_+_17137023 | 4.93 |
ENSDART00000080712
|
slc43a3b
|
solute carrier family 43, member 3b |
| chr12_-_20120702 | 4.85 |
ENSDART00000153387
ENSDART00000158412 ENSDART00000112768 |
ubald1a
|
UBA-like domain containing 1a |
| chr21_+_21811867 | 4.80 |
ENSDART00000185752
|
neu3.4
|
sialidase 3 (membrane sialidase), tandem duplicate 4 |
| chr10_+_8101729 | 4.79 |
ENSDART00000138875
ENSDART00000123447 ENSDART00000185333 |
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr13_-_25303303 | 4.76 |
ENSDART00000133153
|
plaua
|
plasminogen activator, urokinase a |
| chr25_+_19680962 | 4.63 |
ENSDART00000167260
|
si:dkeyp-110c12.3
|
si:dkeyp-110c12.3 |
| chr16_-_30421275 | 4.56 |
ENSDART00000003752
|
cct3
|
chaperonin containing TCP1, subunit 3 (gamma) |
| chr10_-_15910974 | 4.42 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr7_+_24522308 | 4.32 |
ENSDART00000173542
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr7_+_5494565 | 4.29 |
ENSDART00000135271
|
MYADM
|
si:dkeyp-67a8.2 |
| chr3_+_1724941 | 4.03 |
ENSDART00000193402
|
BX321875.2
|
|
| chr23_+_24272421 | 3.92 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr5_+_9428876 | 3.80 |
ENSDART00000081791
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr4_-_49133107 | 3.78 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr2_+_24376373 | 3.70 |
ENSDART00000135323
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr22_-_29922872 | 3.66 |
ENSDART00000020249
|
dusp5
|
dual specificity phosphatase 5 |
| chr12_-_35054354 | 3.59 |
ENSDART00000075351
|
zgc:112285
|
zgc:112285 |
| chr10_+_42169982 | 3.47 |
ENSDART00000190905
|
CU467905.1
|
|
| chr13_-_28688104 | 3.46 |
ENSDART00000133827
|
pcgf6
|
polycomb group ring finger 6 |
| chr21_+_42717424 | 3.39 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr1_+_45754868 | 3.37 |
ENSDART00000084512
|
pkn1a
|
protein kinase N1a |
| chr22_-_3275888 | 3.36 |
ENSDART00000164743
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr11_+_25459697 | 3.28 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr6_+_58915889 | 3.26 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr2_-_37477654 | 3.20 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr19_-_42424599 | 3.13 |
ENSDART00000077042
|
zgc:153441
|
zgc:153441 |
| chr4_+_28997595 | 3.07 |
ENSDART00000133357
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr22_+_9206514 | 3.05 |
ENSDART00000141625
|
si:ch211-156p11.1
|
si:ch211-156p11.1 |
| chr14_+_743346 | 3.04 |
ENSDART00000110511
|
klb
|
klotho beta |
| chr20_+_35279968 | 2.97 |
ENSDART00000168216
ENSDART00000153332 |
fam49a
|
family with sequence similarity 49, member A |
| chr5_-_71765780 | 2.70 |
ENSDART00000011955
|
gpsm1b
|
G protein signaling modulator 1b |
| chr20_+_6590220 | 2.60 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr15_+_2534740 | 2.52 |
ENSDART00000138469
|
cux1b
|
cut-like homeobox 1b |
| chr3_+_28594713 | 2.52 |
ENSDART00000179799
|
sept12
|
septin 12 |
| chr4_+_28998230 | 2.50 |
ENSDART00000183828
|
si:dkey-13e3.1
|
si:dkey-13e3.1 |
| chr3_+_32553714 | 2.42 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr23_+_28378543 | 2.41 |
ENSDART00000145327
|
zgc:153867
|
zgc:153867 |
| chr22_-_382955 | 2.38 |
ENSDART00000082406
|
ora3
|
olfactory receptor class A related 3 |
| chr14_-_10492072 | 2.37 |
ENSDART00000182525
|
lpar4
|
lysophosphatidic acid receptor 4 |
| chr10_-_690072 | 2.35 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr13_+_9496748 | 2.28 |
ENSDART00000147050
ENSDART00000102120 |
CR848040.4
|
|
| chr1_+_50976975 | 2.18 |
ENSDART00000022290
ENSDART00000140982 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr2_+_31804582 | 2.11 |
ENSDART00000086646
|
rnf182
|
ring finger protein 182 |
| chr2_+_47718605 | 2.10 |
ENSDART00000189180
ENSDART00000148824 |
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr4_-_22310956 | 2.09 |
ENSDART00000162585
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr24_-_20192996 | 2.07 |
ENSDART00000011143
|
myd88
|
myeloid differentiation primary response 88 |
| chr11_+_6281647 | 2.03 |
ENSDART00000002459
|
ctns
|
cystinosin, lysosomal cystine transporter |
| chr21_+_4313039 | 1.91 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr18_+_45504362 | 1.88 |
ENSDART00000140089
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr4_+_33461796 | 1.85 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr13_-_50341004 | 1.74 |
ENSDART00000038120
|
cacul1
|
CDK2 associated cullin domain 1 |
| chr21_+_6114709 | 1.73 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr16_-_12496632 | 1.71 |
ENSDART00000019941
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr8_+_26268726 | 1.69 |
ENSDART00000180883
|
slc26a6
|
solute carrier family 26, member 6 |
| chr3_-_33395233 | 1.66 |
ENSDART00000167349
|
si:dkey-283b1.6
|
si:dkey-283b1.6 |
| chr7_-_28549361 | 1.59 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr21_+_30721733 | 1.51 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr13_-_9091354 | 1.45 |
ENSDART00000140319
ENSDART00000142697 |
HTRA2 (1 of many)
|
si:dkey-112g5.13 |
| chr16_-_24832038 | 1.44 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr24_-_7728409 | 1.44 |
ENSDART00000131392
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr13_-_9070754 | 1.38 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr4_+_12612723 | 1.32 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr13_-_377256 | 1.31 |
ENSDART00000137398
|
ch1073-291c23.1
|
ch1073-291c23.1 |
| chr13_-_9045879 | 1.31 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr8_+_25902170 | 1.31 |
ENSDART00000193130
|
rhoab
|
ras homolog gene family, member Ab |
| chr21_+_4388489 | 1.25 |
ENSDART00000144555
|
HTRA2 (1 of many)
|
si:dkey-84o3.3 |
| chr14_-_25309058 | 1.24 |
ENSDART00000159569
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr15_-_39945036 | 1.23 |
ENSDART00000192481
|
msh5
|
mutS homolog 5 |
| chr1_+_44173245 | 1.23 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr4_-_49582758 | 1.19 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr20_+_20186706 | 1.17 |
ENSDART00000002507
|
rhoj
|
ras homolog family member J |
| chr13_+_9468535 | 1.15 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr3_+_54761569 | 1.14 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr3_+_3046909 | 1.09 |
ENSDART00000122435
|
BX004816.2
|
|
| chr1_+_44173506 | 1.00 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr1_+_53714734 | 1.00 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr21_-_25555355 | 0.99 |
ENSDART00000144228
|
si:dkey-17e16.9
|
si:dkey-17e16.9 |
| chr8_+_8937723 | 0.94 |
ENSDART00000145970
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr13_-_9111927 | 0.93 |
ENSDART00000133815
ENSDART00000109783 ENSDART00000142540 ENSDART00000192613 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-19p15.4 si:dkey-112g5.12 |
| chr13_-_9284407 | 0.92 |
ENSDART00000164798
ENSDART00000131956 ENSDART00000140150 |
HTRA2 (1 of many)
|
si:dkey-33c12.14 |
| chr17_+_18031304 | 0.90 |
ENSDART00000127259
|
setd3
|
SET domain containing 3 |
| chr25_-_893464 | 0.82 |
ENSDART00000159321
|
znf609a
|
zinc finger protein 609a |
| chr10_-_32920690 | 0.67 |
ENSDART00000136245
|
cux1a
|
cut-like homeobox 1a |
| chr18_+_50669456 | 0.67 |
ENSDART00000163005
|
si:dkey-151j17.4
|
si:dkey-151j17.4 |
| chr13_+_9521629 | 0.67 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr3_-_40945710 | 0.66 |
ENSDART00000138719
ENSDART00000102416 |
cyp3c4
|
cytochrome P450, family 3, subfamily C, polypeptide 4 |
| chr1_+_8492379 | 0.61 |
ENSDART00000184518
|
myo15ab
|
myosin XVAb |
| chr25_+_17860798 | 0.47 |
ENSDART00000146845
|
pth1a
|
parathyroid hormone 1a |
| chr7_+_71764883 | 0.43 |
ENSDART00000166865
|
myl12.1
|
myosin, light chain 12, genome duplicate 1 |
| chr19_-_20403507 | 0.40 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr20_-_2949028 | 0.34 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr1_+_57331813 | 0.33 |
ENSDART00000152440
ENSDART00000062841 |
epn3b
|
epsin 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of srfa+srfb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.7 | 80.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 10.7 | 32.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 8.0 | 31.9 | GO:0003413 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 7.3 | 43.8 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 6.0 | 30.1 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 5.5 | 22.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 4.7 | 37.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 3.3 | 75.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 2.5 | 34.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 2.1 | 6.2 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 1.7 | 8.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 1.7 | 10.0 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 1.5 | 23.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 1.3 | 5.3 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 1.3 | 34.0 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 1.3 | 44.1 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 1.1 | 6.5 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 1.0 | 2.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.0 | 175.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.9 | 5.6 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.9 | 17.1 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.9 | 19.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.9 | 95.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.9 | 11.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.8 | 3.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.7 | 17.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.7 | 7.7 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.7 | 18.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.6 | 11.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.6 | 29.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.6 | 3.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.5 | 5.1 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.5 | 5.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 4.8 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.5 | 20.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.4 | 2.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.4 | 1.7 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.4 | 137.5 | GO:0061061 | muscle structure development(GO:0061061) |
| 0.4 | 19.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.4 | 4.8 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.4 | 14.0 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.4 | 18.4 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.3 | 9.0 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.3 | 1.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 1.7 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 12.6 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.2 | 3.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 3.0 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.2 | 2.0 | GO:0070293 | renal absorption(GO:0070293) |
| 0.2 | 26.1 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.2 | 1.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 1.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 1.0 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 34.1 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.2 | 13.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 5.5 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 3.9 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 4.6 | GO:2000027 | regulation of organ morphogenesis(GO:2000027) |
| 0.1 | 1.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 1.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 5.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 3.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.3 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 1.0 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 2.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.9 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 11.1 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 2.5 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 21.4 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 2.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 5.5 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 3.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.9 | GO:0007601 | visual perception(GO:0007601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.1 | 144.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 6.8 | 34.2 | GO:0097433 | dense body(GO:0097433) |
| 6.3 | 44.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 4.8 | 99.8 | GO:0031430 | M band(GO:0031430) |
| 4.7 | 37.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 4.4 | 17.8 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 3.4 | 33.8 | GO:0031672 | A band(GO:0031672) |
| 3.4 | 158.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 2.4 | 158.0 | GO:0005884 | actin filament(GO:0005884) |
| 1.9 | 30.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 1.1 | 5.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 1.0 | 122.0 | GO:0016459 | myosin complex(GO:0016459) |
| 1.0 | 32.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.8 | 31.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.8 | 58.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.6 | 11.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.5 | 5.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 19.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.4 | 4.6 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 6.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.4 | 15.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.3 | 82.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.2 | 29.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 23.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 19.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 7.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 20.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 3.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 5.4 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 17.7 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 2.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 14.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 12.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 8.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 34.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 8.8 | 44.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 7.5 | 150.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 6.8 | 34.2 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 4.7 | 37.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 3.5 | 31.2 | GO:0015250 | water channel activity(GO:0015250) |
| 2.5 | 34.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 2.3 | 32.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 2.3 | 33.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 1.7 | 17.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.7 | 10.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.3 | 484.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.9 | 5.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.8 | 5.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.8 | 12.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.7 | 12.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.7 | 2.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.6 | 5.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 6.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.5 | 5.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 4.8 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 2.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.4 | 19.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.4 | 3.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 34.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.3 | 15.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.3 | 3.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 5.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 3.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 7.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 5.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 2.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.9 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 18.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 3.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 34.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 1.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 38.3 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.1 | 6.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 70.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 3.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 2.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 4.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 29.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.9 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 6.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 11.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 1.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 2.1 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 5.6 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 2.5 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.4 | GO:0008134 | transcription factor binding(GO:0008134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 17.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.7 | 71.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.7 | 7.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.7 | 16.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.6 | 5.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.4 | 31.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 10.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 5.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 24.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 7.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 2.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 7.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 63.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 2.2 | 6.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 2.0 | 10.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.9 | 43.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.8 | 37.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 1.1 | 5.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 1.0 | 7.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.9 | 5.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.8 | 34.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 5.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 3.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 2.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.3 | 4.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.3 | 3.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 4.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 1.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.2 | 2.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 3.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 7.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 5.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |