Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
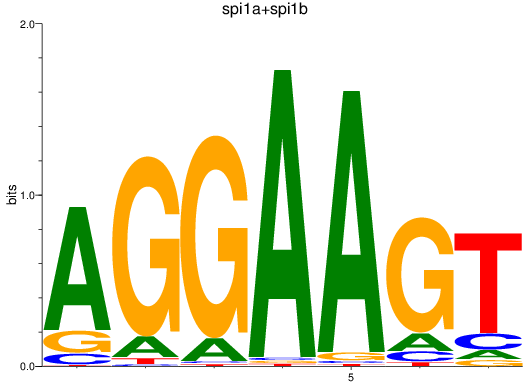
Results for spi1a+spi1b
Z-value: 4.34
Transcription factors associated with spi1a+spi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
spi1b
|
ENSDARG00000000767 | Spi-1 proto-oncogene b |
|
spi1a
|
ENSDARG00000067797 | Spi-1 proto-oncogene a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| spi1a | dr11_v1_chr25_+_35553542_35553542 | 0.78 | 7.8e-21 | Click! |
| spi1b | dr11_v1_chr7_-_32659048_32659048 | 0.75 | 5.8e-18 | Click! |
Activity profile of spi1a+spi1b motif
Sorted Z-values of spi1a+spi1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_43872889 | 40.25 |
ENSDART00000170553
|
mslna
|
mesothelin a |
| chr6_-_8489810 | 38.27 |
ENSDART00000124643
|
rasal3
|
RAS protein activator like 3 |
| chr15_+_20239141 | 37.98 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr21_+_25765734 | 36.97 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr7_-_34265481 | 34.45 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr15_+_12435975 | 34.27 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr3_-_32859335 | 33.00 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr6_-_49063085 | 31.84 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr7_+_49664174 | 31.49 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr23_-_10177442 | 30.33 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr15_-_29598679 | 29.92 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr16_+_17715243 | 29.33 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr13_+_8840772 | 28.31 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr10_+_4987766 | 27.71 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr10_+_13209580 | 27.59 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr13_-_50108337 | 27.53 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr22_+_5478353 | 27.42 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr19_-_40198478 | 26.95 |
ENSDART00000191736
|
grn2
|
granulin 2 |
| chr23_+_26142613 | 26.82 |
ENSDART00000165046
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr20_+_38285671 | 26.72 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr9_+_24088062 | 26.41 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr16_+_48753664 | 25.87 |
ENSDART00000155148
|
si:ch73-31d8.2
|
si:ch73-31d8.2 |
| chr8_+_19356072 | 25.54 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr9_-_443451 | 25.38 |
ENSDART00000165642
|
si:dkey-11f4.14
|
si:dkey-11f4.14 |
| chr16_+_29492749 | 25.23 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr15_+_12436220 | 25.17 |
ENSDART00000169894
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr16_-_31756859 | 24.95 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr22_-_15577060 | 24.93 |
ENSDART00000176291
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr24_-_31306724 | 24.87 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr8_+_32406885 | 24.80 |
ENSDART00000167600
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr5_+_37087583 | 24.74 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr23_+_26142807 | 24.69 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr15_+_46356879 | 24.62 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr1_-_37383539 | 24.56 |
ENSDART00000127579
|
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr7_-_49892991 | 24.55 |
ENSDART00000126240
|
cd44a
|
CD44 molecule (Indian blood group) a |
| chr7_-_2039060 | 24.52 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr3_-_30941362 | 23.27 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr3_+_40809011 | 23.09 |
ENSDART00000033713
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr7_+_34794829 | 22.51 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr13_-_34858500 | 22.42 |
ENSDART00000184843
|
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr21_+_28747069 | 22.16 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr11_-_18384534 | 22.10 |
ENSDART00000156499
|
prkcdb
|
protein kinase C, delta b |
| chr7_+_19482084 | 22.01 |
ENSDART00000173873
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr20_+_10498986 | 21.63 |
ENSDART00000064114
|
zgc:100997
|
zgc:100997 |
| chr12_-_30558694 | 21.62 |
ENSDART00000153417
|
si:ch211-28p3.3
|
si:ch211-28p3.3 |
| chr1_-_45347393 | 21.17 |
ENSDART00000173024
|
si:ch211-243a20.4
|
si:ch211-243a20.4 |
| chr16_-_36099492 | 20.86 |
ENSDART00000180905
|
CU499336.2
|
|
| chr23_+_44599012 | 20.85 |
ENSDART00000125198
|
pfn1
|
profilin 1 |
| chr14_+_29769336 | 20.77 |
ENSDART00000105898
|
si:dkey-34l15.1
|
si:dkey-34l15.1 |
| chr1_-_45157243 | 20.76 |
ENSDART00000131882
|
mucms1
|
mucin, multiple PTS and SEA group, member 1 |
| chr2_+_24762567 | 20.66 |
ENSDART00000078866
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr6_+_112579 | 20.56 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr11_-_21404044 | 20.56 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr23_+_31815423 | 20.50 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr21_+_22985078 | 20.42 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr20_+_26683933 | 19.87 |
ENSDART00000139852
ENSDART00000077751 |
foxq1b
|
forkhead box Q1b |
| chr17_-_8886735 | 19.64 |
ENSDART00000121997
|
nkl.3
|
NK-lysin tandem duplicate 3 |
| chr21_+_15723069 | 19.60 |
ENSDART00000149126
ENSDART00000130628 |
p2rx4a
|
purinergic receptor P2X, ligand-gated ion channel, 4a |
| chr3_+_29942338 | 19.51 |
ENSDART00000158220
|
ifi35
|
interferon-induced protein 35 |
| chr9_+_30387930 | 19.43 |
ENSDART00000112827
|
si:dkey-18p12.4
|
si:dkey-18p12.4 |
| chr3_+_18579806 | 19.33 |
ENSDART00000180967
ENSDART00000089765 |
arhgap17b
|
Rho GTPase activating protein 17b |
| chr17_-_2039511 | 19.14 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr16_-_41990421 | 19.05 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr7_-_26603743 | 18.95 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr7_-_5316901 | 18.95 |
ENSDART00000181505
ENSDART00000124367 |
si:cabz01074946.1
|
si:cabz01074946.1 |
| chr3_+_36275633 | 18.78 |
ENSDART00000185027
ENSDART00000149532 ENSDART00000102883 ENSDART00000148444 |
zgc:86896
|
zgc:86896 |
| chr20_+_38276690 | 18.76 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr20_-_19864131 | 18.75 |
ENSDART00000057819
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr3_-_53508580 | 18.48 |
ENSDART00000073978
|
zgc:171711
|
zgc:171711 |
| chr14_+_20911310 | 18.28 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr16_+_38201840 | 18.23 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr16_+_21789703 | 17.93 |
ENSDART00000153617
|
trim108
|
tripartite motif containing 108 |
| chr19_-_325584 | 17.81 |
ENSDART00000134266
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr7_-_7810348 | 17.77 |
ENSDART00000171984
|
cxcl19
|
chemokine (C-X-C motif) ligand 19 |
| chr1_+_40308077 | 17.52 |
ENSDART00000138992
|
vwa10.2
|
von Willebrand factor A domain containing 10, tandem duplicate 2 |
| chr1_-_37383741 | 17.43 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr7_+_15736230 | 17.27 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr19_-_34927201 | 17.26 |
ENSDART00000076518
|
sla1
|
Src-like-adaptor 1 |
| chr20_+_46040666 | 17.24 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr16_+_29492937 | 17.14 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr15_-_29598444 | 17.14 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr16_-_24832038 | 16.86 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr10_-_41980797 | 16.86 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr3_-_27647845 | 16.68 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr4_-_992063 | 16.68 |
ENSDART00000181630
ENSDART00000183898 ENSDART00000160902 |
naga
|
N-acetylgalactosaminidase, alpha |
| chr21_+_28747236 | 16.59 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr7_+_19483277 | 16.56 |
ENSDART00000173750
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr12_+_22576404 | 16.52 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr21_+_30937690 | 16.49 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr25_+_4750972 | 16.42 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr2_-_37744951 | 16.36 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr5_-_37959874 | 16.31 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr15_-_2652640 | 16.15 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr19_-_24958393 | 16.13 |
ENSDART00000098592
|
si:ch211-195b13.6
|
si:ch211-195b13.6 |
| chr9_-_56272465 | 16.06 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr17_-_5860222 | 15.99 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr10_+_38417512 | 15.95 |
ENSDART00000112457
|
samsn1b
|
SAM domain, SH3 domain and nuclear localisation signals 1b |
| chr20_+_9223514 | 15.91 |
ENSDART00000023293
|
kcnk5b
|
potassium channel, subfamily K, member 5b |
| chr18_+_22109379 | 15.90 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr3_+_29510818 | 15.85 |
ENSDART00000055407
ENSDART00000193743 ENSDART00000123619 |
rac2
|
Rac family small GTPase 2 |
| chr15_+_36445350 | 15.80 |
ENSDART00000154552
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr3_+_24641489 | 15.75 |
ENSDART00000055590
ENSDART00000186635 |
zgc:113411
|
zgc:113411 |
| chr24_-_31904924 | 15.74 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr3_+_2971852 | 15.72 |
ENSDART00000059271
|
BX004816.1
|
|
| chr17_-_6508406 | 15.35 |
ENSDART00000002778
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr10_+_26612321 | 15.19 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr11_+_44135351 | 15.18 |
ENSDART00000182914
|
FO704721.1
|
|
| chr20_-_7293837 | 15.18 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr7_+_29115890 | 15.18 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr22_-_3914162 | 15.09 |
ENSDART00000187174
ENSDART00000190612 ENSDART00000187928 ENSDART00000057224 ENSDART00000184758 |
mhc1uma
|
major histocompatibility complex class I UMA |
| chr10_-_22127942 | 15.05 |
ENSDART00000133374
|
ponzr2
|
plac8 onzin related protein 2 |
| chr14_-_40797117 | 15.03 |
ENSDART00000122369
|
elf1
|
E74-like ETS transcription factor 1 |
| chr15_-_18209672 | 14.98 |
ENSDART00000141508
ENSDART00000136280 |
btr16
|
bloodthirsty-related gene family, member 16 |
| chr9_+_41224100 | 14.96 |
ENSDART00000141792
|
stat1b
|
signal transducer and activator of transcription 1b |
| chr10_-_20650302 | 14.91 |
ENSDART00000142028
ENSDART00000064662 |
rassf2b
|
Ras association (RalGDS/AF-6) domain family member 2b |
| chr22_+_3914318 | 14.86 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr10_-_7785930 | 14.86 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr21_+_27448856 | 14.80 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr5_-_69948099 | 14.72 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr8_-_3312384 | 14.67 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr15_+_5088210 | 14.65 |
ENSDART00000183423
|
mxf
|
myxovirus (influenza virus) resistance F |
| chr8_-_28349859 | 14.63 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr5_-_25582721 | 14.41 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr10_-_35149513 | 14.40 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr5_-_23909934 | 14.39 |
ENSDART00000142516
|
si:ch211-135f11.1
|
si:ch211-135f11.1 |
| chr12_-_30540699 | 14.36 |
ENSDART00000167712
ENSDART00000102464 |
zgc:153920
|
zgc:153920 |
| chr14_+_35428152 | 14.34 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr7_+_39418869 | 14.32 |
ENSDART00000169195
|
CT030188.1
|
|
| chr10_+_8875195 | 14.25 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr4_+_7677318 | 14.21 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr8_-_12867128 | 14.00 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr4_-_22310956 | 13.90 |
ENSDART00000162585
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr2_-_6548838 | 13.88 |
ENSDART00000052419
|
rgs18
|
regulator of G protein signaling 18 |
| chr21_-_25295087 | 13.85 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr9_+_45839260 | 13.83 |
ENSDART00000114814
|
twist2
|
twist2 |
| chr5_-_68876211 | 13.78 |
ENSDART00000097254
|
sftpbb
|
surfactant protein Bb |
| chr10_-_24648228 | 13.75 |
ENSDART00000081834
ENSDART00000132830 |
stoml3b
|
stomatin (EPB72)-like 3b |
| chr7_+_22657566 | 13.67 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr6_-_40657653 | 13.66 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr25_+_34014523 | 13.55 |
ENSDART00000182856
|
anxa2a
|
annexin A2a |
| chr13_-_32898962 | 13.49 |
ENSDART00000163757
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr15_+_36457888 | 13.48 |
ENSDART00000155100
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr5_-_61445697 | 13.44 |
ENSDART00000050898
|
ncf1
|
neutrophil cytosolic factor 1 |
| chr3_+_32492467 | 13.42 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr24_-_32582378 | 13.40 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr19_+_40350468 | 13.34 |
ENSDART00000087444
|
hepacam2
|
HEPACAM family member 2 |
| chr12_-_4243268 | 13.26 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr14_+_23709543 | 13.22 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr1_-_25177086 | 13.21 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr19_-_25005609 | 13.20 |
ENSDART00000151129
|
xkr8.2
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 2 |
| chr22_-_15717897 | 13.14 |
ENSDART00000008424
|
malt2
|
MALT paracaspase 2 |
| chr8_-_36469117 | 13.11 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr24_-_25096199 | 13.09 |
ENSDART00000185076
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr8_-_19975087 | 13.05 |
ENSDART00000182220
|
lpxn
|
leupaxin |
| chr20_-_13660600 | 13.01 |
ENSDART00000063826
|
TAGAP
|
si:ch211-122h15.4 |
| chr22_+_5687615 | 12.99 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr7_+_26029672 | 12.98 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr14_-_16807206 | 12.96 |
ENSDART00000157957
|
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr16_+_20934353 | 12.93 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr7_+_1045637 | 12.90 |
ENSDART00000111531
|
epdl1
|
ependymin-like 1 |
| chr21_-_40174647 | 12.90 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr10_-_24391716 | 12.88 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr2_-_55337585 | 12.88 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr3_-_40528333 | 12.85 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr9_+_23772516 | 12.66 |
ENSDART00000183126
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr1_+_54834119 | 12.65 |
ENSDART00000140020
|
si:ch211-196h16.4
|
si:ch211-196h16.4 |
| chr19_+_43523690 | 12.62 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr7_-_60351537 | 12.61 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr10_-_28395620 | 12.54 |
ENSDART00000168907
|
CR392341.4
|
|
| chr25_+_18475032 | 12.50 |
ENSDART00000073564
|
tes
|
testis derived transcript (3 LIM domains) |
| chr19_+_18758014 | 12.50 |
ENSDART00000158935
|
clic1
|
chloride intracellular channel 1 |
| chr20_-_35470891 | 12.50 |
ENSDART00000152993
ENSDART00000016090 |
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr3_-_50136424 | 12.48 |
ENSDART00000188843
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr16_-_25233515 | 12.47 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr6_-_40842768 | 12.46 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr21_-_26114886 | 12.38 |
ENSDART00000139320
|
nipal4
|
NIPA-like domain containing 4 |
| chr20_-_34127415 | 12.35 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr12_-_22400999 | 12.31 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr3_+_42923275 | 12.29 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr12_-_28983584 | 12.28 |
ENSDART00000112374
|
zgc:171713
|
zgc:171713 |
| chr5_+_28271412 | 12.24 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr19_-_5669122 | 12.23 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr12_-_9438227 | 12.21 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr11_+_29975830 | 12.20 |
ENSDART00000148929
|
si:ch73-226l13.2
|
si:ch73-226l13.2 |
| chr5_+_20421539 | 12.19 |
ENSDART00000164499
|
selplg
|
selectin P ligand |
| chr6_-_42949184 | 12.19 |
ENSDART00000147208
|
edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr25_+_13191391 | 12.16 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr7_+_19482877 | 12.11 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr9_+_21165017 | 12.10 |
ENSDART00000145933
ENSDART00000142985 |
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr12_-_15205087 | 12.08 |
ENSDART00000010068
|
sult1st6
|
sulfotransferase family 1, cytosolic sulfotransferase 6 |
| chr5_-_26879302 | 12.05 |
ENSDART00000098571
ENSDART00000139086 |
zgc:64051
|
zgc:64051 |
| chr20_+_10498704 | 12.05 |
ENSDART00000192985
|
zgc:100997
|
zgc:100997 |
| chr11_+_1602916 | 12.04 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr24_-_21131607 | 12.02 |
ENSDART00000010126
|
zdhhc23b
|
zinc finger, DHHC-type containing 23b |
| chr9_+_13120419 | 11.92 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr7_+_35268054 | 11.86 |
ENSDART00000113842
|
dpep2
|
dipeptidase 2 |
| chr22_-_17688868 | 11.86 |
ENSDART00000012336
ENSDART00000147070 |
tjp3
|
tight junction protein 3 |
| chr9_-_25993849 | 11.79 |
ENSDART00000144840
|
arhgap15
|
Rho GTPase activating protein 15 |
| chr21_-_35534401 | 11.77 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr18_+_15937610 | 11.76 |
ENSDART00000061134
ENSDART00000154505 |
itpr2
|
inositol 1,4,5-trisphosphate receptor, type 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of spi1a+spi1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.2 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 7.0 | 21.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 6.9 | 20.7 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 6.4 | 38.4 | GO:0070254 | mucus secretion(GO:0070254) |
| 6.1 | 18.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 5.8 | 17.4 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 5.0 | 14.9 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 4.9 | 29.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 4.7 | 18.7 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 4.5 | 13.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 4.5 | 17.8 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 4.3 | 17.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 4.3 | 17.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 3.8 | 11.4 | GO:0002855 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 3.5 | 14.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 3.5 | 20.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 3.4 | 37.9 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 3.4 | 20.5 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 3.2 | 16.2 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 3.2 | 22.3 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 3.2 | 28.6 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 3.1 | 34.4 | GO:0045453 | bone resorption(GO:0045453) |
| 3.1 | 15.6 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 3.1 | 12.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 3.1 | 12.2 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 3.0 | 15.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 2.9 | 8.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 2.9 | 20.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 2.9 | 60.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 2.8 | 11.3 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 2.8 | 8.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 2.7 | 18.6 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 2.6 | 28.9 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 2.6 | 17.9 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 2.5 | 5.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 2.5 | 12.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 2.4 | 14.4 | GO:0032663 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 2.3 | 7.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 2.3 | 11.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 2.3 | 9.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 2.2 | 31.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 2.2 | 8.8 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 2.2 | 8.8 | GO:0050851 | antigen receptor-mediated signaling pathway(GO:0050851) B cell receptor signaling pathway(GO:0050853) |
| 2.2 | 8.8 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 2.2 | 6.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) anterior lateral line neuromast hair cell development(GO:0035676) |
| 2.2 | 49.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 2.2 | 6.5 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 2.1 | 25.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 2.0 | 12.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 2.0 | 12.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 2.0 | 8.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 2.0 | 11.7 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 1.9 | 9.5 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 1.9 | 5.7 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 1.9 | 32.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 1.9 | 7.5 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 1.9 | 15.0 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 1.9 | 9.3 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 1.9 | 5.6 | GO:0046048 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 1.8 | 9.2 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 1.8 | 3.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 1.7 | 12.1 | GO:0051705 | multi-organism behavior(GO:0051705) |
| 1.7 | 5.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 1.7 | 6.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 1.7 | 6.8 | GO:2000383 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 1.6 | 24.7 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 1.6 | 6.6 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 1.6 | 19.6 | GO:0033198 | response to ATP(GO:0033198) |
| 1.6 | 21.0 | GO:0007172 | signal complex assembly(GO:0007172) |
| 1.6 | 4.8 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 1.6 | 8.1 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 1.6 | 29.9 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 1.6 | 26.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 1.6 | 4.7 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 1.5 | 7.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 1.5 | 4.6 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 1.5 | 7.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 1.5 | 9.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 1.5 | 3.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 1.5 | 5.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.5 | 7.4 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 1.5 | 14.7 | GO:0051125 | regulation of actin nucleation(GO:0051125) |
| 1.5 | 4.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 1.5 | 1.5 | GO:0001818 | negative regulation of cytokine production(GO:0001818) |
| 1.4 | 8.7 | GO:0010269 | response to selenium ion(GO:0010269) |
| 1.4 | 12.9 | GO:0042113 | B cell activation(GO:0042113) |
| 1.4 | 15.4 | GO:1901072 | chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 1.4 | 79.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 1.4 | 20.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 1.4 | 11.0 | GO:0036372 | opsin transport(GO:0036372) |
| 1.3 | 29.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 1.3 | 8.0 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 1.3 | 11.9 | GO:0030104 | water homeostasis(GO:0030104) |
| 1.3 | 7.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.3 | 14.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.3 | 6.4 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 1.2 | 24.6 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 1.2 | 4.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 1.2 | 52.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 1.2 | 15.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 1.2 | 8.1 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 1.2 | 15.0 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 1.1 | 6.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.1 | 9.2 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 1.1 | 4.6 | GO:0002637 | regulation of immunoglobulin production(GO:0002637) positive regulation of immunoglobulin production(GO:0002639) |
| 1.1 | 10.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 1.1 | 12.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 1.1 | 3.3 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 1.1 | 23.3 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 1.1 | 7.8 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 1.1 | 25.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.1 | 15.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 1.1 | 7.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 1.1 | 2.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 1.1 | 23.4 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 1.1 | 11.7 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 1.1 | 6.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 1.0 | 4.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.0 | 4.1 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 1.0 | 2.1 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 1.0 | 5.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 1.0 | 9.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.0 | 9.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 1.0 | 2.0 | GO:0060306 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 1.0 | 9.0 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 1.0 | 3.0 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 1.0 | 9.8 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.0 | 19.6 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 1.0 | 74.2 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 1.0 | 9.7 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 1.0 | 5.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 1.0 | 5.8 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.9 | 10.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.9 | 3.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.9 | 6.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.9 | 4.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.9 | 4.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.9 | 9.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.9 | 7.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.9 | 28.0 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.9 | 22.3 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.9 | 4.6 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.9 | 13.0 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.9 | 12.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.9 | 2.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.9 | 4.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.9 | 3.7 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.9 | 10.9 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.9 | 26.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.9 | 4.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.9 | 23.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.9 | 11.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.9 | 2.6 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.9 | 3.5 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.9 | 3.4 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.8 | 20.7 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.8 | 2.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.8 | 23.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.8 | 2.5 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.8 | 5.7 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.8 | 12.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.8 | 11.3 | GO:0090481 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.8 | 4.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.8 | 14.3 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.8 | 13.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.8 | 24.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.8 | 10.3 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.8 | 32.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.8 | 7.0 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.8 | 14.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.8 | 98.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.8 | 2.3 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.8 | 6.8 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.8 | 6.1 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.8 | 3.8 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.8 | 6.0 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.7 | 7.4 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.7 | 5.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.7 | 16.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.7 | 2.9 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.7 | 1.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.7 | 26.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.7 | 5.7 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.7 | 9.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.7 | 4.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.7 | 8.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.7 | 11.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.7 | 2.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.7 | 4.8 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.7 | 4.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.7 | 14.9 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.7 | 54.7 | GO:0060348 | bone development(GO:0060348) |
| 0.7 | 30.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.7 | 2.0 | GO:0034103 | regulation of tissue remodeling(GO:0034103) regulation of bone remodeling(GO:0046850) |
| 0.6 | 7.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.6 | 1.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.6 | 14.7 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.6 | 3.8 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.6 | 4.4 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.6 | 5.0 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.6 | 6.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.6 | 3.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.6 | 29.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.6 | 1.9 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.6 | 2.5 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.6 | 4.9 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.6 | 26.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.6 | 15.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.6 | 7.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.6 | 4.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.6 | 3.0 | GO:0030828 | nitric oxide mediated signal transduction(GO:0007263) regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.6 | 2.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.6 | 3.6 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.6 | 2.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.6 | 2.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.6 | 3.9 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.5 | 2.7 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.5 | 3.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.5 | 1.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.5 | 2.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.5 | 13.3 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.5 | 1.6 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.5 | 6.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 2.1 | GO:0043096 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) |
| 0.5 | 4.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.5 | 3.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.5 | 7.6 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.5 | 5.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.5 | 38.5 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.5 | 3.9 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.5 | 42.9 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.5 | 2.4 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.5 | 7.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.5 | 1.9 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.5 | 10.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.5 | 7.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.5 | 3.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.5 | 42.6 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.5 | 2.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.5 | 1.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.4 | 4.9 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.4 | 3.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.4 | 12.2 | GO:0010632 | regulation of epithelial cell migration(GO:0010632) |
| 0.4 | 21.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.4 | 1.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.4 | 7.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.4 | 20.8 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.4 | 7.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.4 | 8.6 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.4 | 2.8 | GO:0032475 | otolith formation(GO:0032475) |
| 0.4 | 12.1 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.4 | 6.1 | GO:0061036 | positive regulation of chondrocyte differentiation(GO:0032332) positive regulation of cartilage development(GO:0061036) |
| 0.4 | 5.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.4 | 7.2 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.4 | 6.8 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.4 | 3.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.4 | 11.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.4 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.4 | 5.3 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.4 | 1.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.4 | 0.4 | GO:0045058 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) positive T cell selection(GO:0043368) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T cell selection(GO:0045058) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.4 | 10.9 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.4 | 4.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.4 | 11.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.4 | 11.3 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.4 | 17.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.4 | 4.4 | GO:0051604 | protein maturation(GO:0051604) |
| 0.4 | 5.5 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.4 | 30.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.4 | 1.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 6.9 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.4 | 15.8 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.4 | 14.9 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.4 | 2.1 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.4 | 4.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.4 | 11.2 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.3 | 3.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.3 | 27.1 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.3 | 5.4 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.3 | 8.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.3 | 9.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.3 | 8.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.3 | 6.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.3 | 4.7 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 3.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.3 | 98.7 | GO:0006955 | immune response(GO:0006955) |
| 0.3 | 4.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.3 | 4.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.3 | 1.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.3 | 1.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 10.5 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.3 | 60.3 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.3 | 2.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.3 | 9.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.3 | 1.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 7.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 72.1 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.3 | 3.7 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.3 | 2.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.3 | 1.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.3 | 8.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.3 | 1.1 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.3 | 4.5 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.3 | 1.6 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.3 | 4.0 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.3 | 4.8 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.3 | 2.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.3 | 7.3 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.3 | 1.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 3.1 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.3 | 2.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.3 | 10.3 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.3 | 18.1 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.3 | 16.6 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.2 | 3.5 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 2.5 | GO:0071392 | intracellular estrogen receptor signaling pathway(GO:0030520) cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 6.7 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.2 | 5.9 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.2 | 4.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 11.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.2 | 6.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.2 | 4.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 1.6 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.2 | 2.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 4.0 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.2 | 1.7 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 1.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 1.2 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.2 | 1.6 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.2 | 6.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 1.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 1.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 12.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 2.4 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 24.3 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.2 | 17.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.2 | 6.3 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.2 | 0.7 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.2 | 0.4 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.2 | 2.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 0.5 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.2 | 1.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 4.9 | GO:0098542 | defense response to other organism(GO:0098542) |
| 0.2 | 0.9 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.2 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.2 | 4.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 5.2 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.2 | 1.4 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.2 | 1.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 2.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 4.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.2 | 1.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 2.0 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.2 | 8.7 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.2 | 6.8 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.2 | 7.3 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.2 | 10.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 1.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 6.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 5.3 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) primary miRNA processing(GO:0031053) |
| 0.1 | 0.4 | GO:1903173 | fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 5.5 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 3.7 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 7.4 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 1.8 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.1 | 7.3 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.1 | 1.6 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.1 | 1.6 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 5.6 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
| 0.1 | 4.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 2.5 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.1 | 2.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 5.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 2.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 7.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 40.8 | GO:0006412 | translation(GO:0006412) peptide biosynthetic process(GO:0043043) |
| 0.1 | 1.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.5 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 7.1 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.1 | 1.7 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.6 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 13.7 | GO:0072594 | establishment of protein localization to organelle(GO:0072594) |
| 0.1 | 1.6 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.1 | 5.5 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 1.2 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 1.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.5 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 2.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 0.2 | GO:0032988 | spliceosomal complex disassembly(GO:0000390) ribonucleoprotein complex disassembly(GO:0032988) |
| 0.1 | 1.1 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 0.2 | GO:0046133 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.1 | 1.0 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 2.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.6 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 0.9 | GO:0007044 | cell-substrate junction assembly(GO:0007044) hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 1.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 3.0 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.1 | 2.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 1.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 4.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.8 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 2.0 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.7 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.5 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 35.4 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.5 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) stress granule assembly(GO:0034063) |
| 0.0 | 2.1 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.6 | GO:0046460 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.3 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0042417 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) dopamine metabolic process(GO:0042417) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 27.1 | GO:0097433 | dense body(GO:0097433) |
| 4.8 | 19.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 3.8 | 22.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 3.7 | 11.1 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 3.6 | 14.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 3.4 | 10.2 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 3.4 | 60.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 3.0 | 3.0 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 3.0 | 32.7 | GO:0045095 | keratin filament(GO:0045095) |
| 2.6 | 18.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 2.2 | 17.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 2.2 | 22.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 2.1 | 6.2 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 1.9 | 5.8 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 1.9 | 89.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 1.8 | 25.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 1.6 | 6.6 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 1.6 | 13.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.6 | 6.5 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 1.6 | 23.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 1.5 | 9.3 | GO:0042611 | MHC protein complex(GO:0042611) |
| 1.5 | 4.6 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 1.5 | 10.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 1.5 | 10.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.5 | 8.7 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 1.4 | 73.6 | GO:0005902 | microvillus(GO:0005902) |
| 1.4 | 5.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.3 | 2.7 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 1.3 | 8.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 1.3 | 7.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.3 | 3.9 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 1.3 | 13.0 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 1.3 | 1.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 1.3 | 7.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 1.2 | 6.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.2 | 3.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.1 | 68.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 1.1 | 3.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 1.0 | 21.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 1.0 | 6.2 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 1.0 | 15.2 | GO:0030057 | desmosome(GO:0030057) |
| 1.0 | 2.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 1.0 | 3.9 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.9 | 257.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.9 | 12.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.9 | 6.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.9 | 11.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.8 | 3.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.8 | 16.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.8 | 19.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.8 | 14.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.7 | 5.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.7 | 2.9 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.7 | 4.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.6 | 1.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.6 | 1.9 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.6 | 52.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.6 | 3.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.6 | 73.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.6 | 7.8 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.6 | 4.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.6 | 43.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.6 | 7.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.6 | 65.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.6 | 9.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.6 | 11.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.6 | 15.6 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.6 | 4.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.5 | 1.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.5 | 30.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.5 | 31.1 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.5 | 2.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.5 | 4.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.5 | 7.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.5 | 6.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.5 | 1.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.5 | 10.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.5 | 59.6 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.5 | 4.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 1.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.5 | 6.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.5 | 10.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.5 | 3.7 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.5 | 11.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 2.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.5 | 17.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.4 | 21.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.4 | 17.8 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.4 | 5.8 | GO:0005903 | brush border(GO:0005903) |
| 0.4 | 103.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.4 | 2.0 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.4 | 0.4 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.4 | 1.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.4 | 10.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.4 | 5.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.4 | 1.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 1.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 5.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 26.2 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.3 | 1.6 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.3 | 13.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.3 | 1.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 3.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 3.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.3 | 11.0 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.3 | 5.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 1.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 1.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 1.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.3 | 26.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 4.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 1.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.3 | 3.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 1.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 3.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 2.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.2 | 23.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 20.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 3.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 1.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 8.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 0.7 | GO:0000323 | lytic vacuole(GO:0000323) |
| 0.2 | 1.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 27.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 11.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 2.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 25.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.2 | 1.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 9.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.2 | 2.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 1.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 2.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 15.2 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.2 | 3.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 5.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 2.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 6.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 8.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 11.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 4.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.2 | 21.6 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.2 | 5.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.6 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 5.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 4.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.3 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 5.2 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 24.8 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.1 | 8.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 5.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 7.7 | GO:0030880 | RNA polymerase complex(GO:0030880) |
| 0.1 | 1.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 5.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 7.5 | GO:0098562 | cytoplasmic side of membrane(GO:0098562) |
| 0.1 | 24.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 2.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 14.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 4.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 143.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 6.2 | GO:0030054 | cell junction(GO:0030054) |
| 0.1 | 1.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 1.2 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 15.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 5.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 5.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 2.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 4.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 5.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.9 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 18.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.1 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 78.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 8.8 | GO:0005886 | plasma membrane(GO:0005886) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 51.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 5.6 | 16.7 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 5.5 | 38.8 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 5.4 | 27.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 5.3 | 16.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 5.2 | 20.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 4.6 | 18.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 4.5 | 17.8 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 4.4 | 13.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 4.4 | 17.6 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 4.1 | 28.9 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 4.0 | 12.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 3.5 | 24.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 3.4 | 10.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 3.2 | 9.6 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 3.1 | 9.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 3.1 | 12.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 3.0 | 9.1 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 3.0 | 27.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 2.8 | 19.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 2.7 | 13.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 2.6 | 7.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 2.5 | 25.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 2.5 | 15.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 2.5 | 7.5 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 2.4 | 14.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 2.4 | 9.5 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 2.4 | 11.9 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 2.4 | 21.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 2.3 | 21.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 2.2 | 6.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 2.2 | 23.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.1 | 29.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 2.1 | 24.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 2.0 | 8.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.9 | 38.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 1.9 | 13.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 1.9 | 5.8 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 1.9 | 11.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 1.9 | 7.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 1.9 | 5.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.9 | 5.6 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 1.8 | 18.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 1.8 | 5.5 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 1.7 | 7.0 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 1.7 | 6.9 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 1.7 | 15.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.7 | 13.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 1.7 | 6.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 1.6 | 19.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.6 | 16.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 1.5 | 7.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 1.5 | 15.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.5 | 7.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 1.5 | 19.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 1.5 | 31.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 1.4 | 11.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 1.4 | 15.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 1.4 | 12.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.4 | 17.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 1.3 | 9.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 1.3 | 24.8 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 1.3 | 13.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 1.3 | 9.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.3 | 3.8 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 1.3 | 3.8 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.3 | 10.1 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.2 | 56.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 1.2 | 3.7 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 1.2 | 3.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.2 | 9.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 1.2 | 8.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 1.1 | 11.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 1.1 | 24.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.1 | 12.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 1.1 | 74.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 1.1 | 3.3 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 1.1 | 12.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 1.1 | 59.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 1.1 | 12.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.1 | 5.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.1 | 22.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 1.1 | 7.5 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.1 | 4.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 1.1 | 9.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.1 | 6.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 1.0 | 37.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 1.0 | 11.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.0 | 5.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 1.0 | 4.8 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.9 | 21.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.9 | 4.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.9 | 3.7 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.9 | 13.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.9 | 13.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.9 | 4.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.9 | 3.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.9 | 20.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.9 | 5.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.9 | 5.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.8 | 4.2 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.8 | 2.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.8 | 20.7 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.8 | 8.3 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.8 | 12.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.8 | 2.5 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.8 | 5.7 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.8 | 64.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.8 | 17.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.8 | 158.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.8 | 8.0 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.8 | 4.8 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.8 | 3.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.8 | 3.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.8 | 3.8 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.8 | 5.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.8 | 5.3 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.8 | 3.0 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.7 | 11.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.7 | 11.5 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.7 | 7.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.7 | 1.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.7 | 2.1 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.7 | 2.0 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.7 | 9.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.7 | 17.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.6 | 22.8 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.6 | 4.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.6 | 3.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.6 | 1.9 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.6 | 8.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.6 | 1.9 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.6 | 2.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.6 | 1.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.6 | 84.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.6 | 10.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.6 | 2.9 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.6 | 28.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.6 | 3.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.6 | 5.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.6 | 1.7 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.6 | 7.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.6 | 2.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.6 | 27.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.6 | 5.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.5 | 2.7 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.5 | 6.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.5 | 2.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.5 | 5.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.5 | 3.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 24.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.5 | 20.1 | GO:0019956 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.5 | 2.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.5 | 2.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 9.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.5 | 10.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.5 | 4.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.5 | 3.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 6.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.5 | 5.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.5 | 6.0 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.5 | 2.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.5 | 57.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 1.9 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.5 | 6.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.5 | 12.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.5 | 1.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.5 | 5.9 | GO:0070035 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.4 | 9.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.4 | 4.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 24.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.4 | 3.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 8.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.4 | 1.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 131.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.4 | 4.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 9.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.4 | 4.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.4 | 3.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.4 | 83.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.4 | 7.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.4 | 7.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 5.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 4.7 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.4 | 1.9 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.4 | 2.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 10.2 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.4 | 144.7 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.4 | 1.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 12.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.4 | 2.9 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.4 | 4.0 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 7.6 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.4 | 1.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.4 | 18.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.4 | 2.5 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.4 | 3.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 8.7 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.3 | 6.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 7.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 6.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.3 | 22.4 | GO:0032934 | sterol binding(GO:0032934) |
| 0.3 | 1.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 2.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 2.9 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.3 | 3.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.3 | 2.2 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.3 | 3.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 3.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.5 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.3 | 8.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 4.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 22.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.3 | 26.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.3 | 3.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.3 | 2.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.3 | 1.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.3 | 0.8 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.3 | 1.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 2.0 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.2 | 5.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 3.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 0.7 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.2 | 3.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 32.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.2 | 3.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 9.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.2 | 2.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 0.7 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.2 | 14.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 1.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 3.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 122.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.2 | 6.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 3.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 4.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 0.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.5 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.2 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 10.9 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.2 | 4.7 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.2 | 1.0 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.2 | 8.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 1.0 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 1.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 4.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 14.7 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.2 | 1.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 2.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 4.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 3.0 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 5.4 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.1 | 0.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 4.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 3.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 1.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 4.0 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.1 | 4.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 11.6 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 1.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 3.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 4.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 6.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.6 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 1.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 1.9 | GO:0016896 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.1 | 2.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 2.0 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.1 | 4.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 60.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 6.4 | GO:0043130 | ubiquitin-like protein binding(GO:0032182) ubiquitin binding(GO:0043130) |
| 0.1 | 5.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.7 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 2.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 15.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 12.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 4.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 3.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 11.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 2.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 5.1 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 7.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 6.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 16.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 8.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.4 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 20.8 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 16.7 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 2.7 | 8.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 2.3 | 18.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 1.7 | 86.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 1.7 | 64.3 | PID EPO PATHWAY | EPO signaling pathway |
| 1.6 | 29.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 1.6 | 17.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 1.6 | 35.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 1.5 | 8.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 1.5 | 29.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 1.4 | 24.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 1.3 | 18.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 1.3 | 24.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.2 | 25.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 1.2 | 9.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 1.2 | 26.7 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 1.2 | 18.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 1.1 | 4.4 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 1.1 | 12.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 1.1 | 9.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 1.1 | 7.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.0 | 14.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.0 | 15.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 1.0 | 22.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.9 | 3.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.9 | 17.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.8 | 35.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.8 | 13.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.8 | 5.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.7 | 25.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.7 | 4.3 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.7 | 9.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.7 | 3.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.6 | 6.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.6 | 16.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.6 | 8.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.6 | 15.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.5 | 7.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 10.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.5 | 10.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.4 | 12.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.4 | 13.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 6.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.4 | 1.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.4 | 1.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.4 | 3.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 6.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 3.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.3 | 4.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 2.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.3 | 4.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.3 | 8.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.3 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.3 | 23.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.3 | 21.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 2.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 3.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 35.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 7.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 3.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 1.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 7.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 22.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.1 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 27.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 4.2 | 50.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 3.8 | 19.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 3.5 | 24.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 3.1 | 34.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 2.8 | 30.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 2.8 | 19.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 2.7 | 21.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 2.6 | 23.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 2.3 | 18.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 2.2 | 12.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 1.9 | 3.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 1.9 | 28.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 1.8 | 94.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 1.7 | 17.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.6 | 12.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 1.5 | 11.8 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.5 | 2.9 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 1.4 | 5.8 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 1.3 | 26.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 1.3 | 2.6 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 1.1 | 10.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 1.1 | 4.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 1.1 | 11.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 1.0 | 29.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 1.0 | 72.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 1.0 | 6.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.0 | 1.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 1.0 | 3.8 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.9 | 19.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.9 | 8.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.9 | 2.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.9 | 8.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.9 | 10.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.8 | 8.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.8 | 16.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.8 | 28.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.8 | 85.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.8 | 6.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.8 | 26.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.7 | 8.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.7 | 10.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.7 | 8.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.7 | 7.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.7 | 16.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.7 | 8.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.6 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.6 | 3.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.6 | 4.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 8.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.6 | 10.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.5 | 13.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.5 | 5.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 8.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.5 | 10.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.5 | 6.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.5 | 12.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.5 | 5.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.4 | 7.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 7.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.4 | 3.6 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.4 | 4.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.4 | 4.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 28.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.4 | 3.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.4 | 1.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 5.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.3 | 3.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 21.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 4.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.3 | 6.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.3 | 4.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 3.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.3 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 12.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.3 | 4.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.3 | 3.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 5.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.2 | 1.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 2.0 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.2 | 7.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.2 | 3.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 3.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 9.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 1.7 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.2 | 3.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 8.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.2 | 4.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.2 | 1.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 8.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 6.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 3.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.2 | 2.3 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.1 | 1.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 3.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 5.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.5 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.1 | REACTOME S PHASE | Genes involved in S Phase |
| 0.1 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.0 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.5 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |