Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
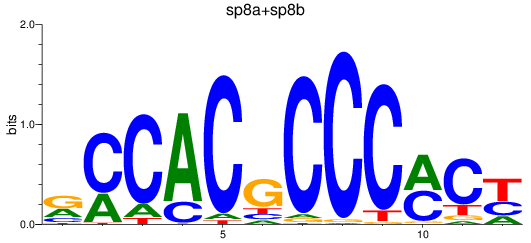
Results for sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
Z-value: 2.09
Transcription factors associated with sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp8a
|
ENSDARG00000011870 | sp8 transcription factor a |
|
sp8b
|
ENSDARG00000056666 | sp8 transcription factor b |
|
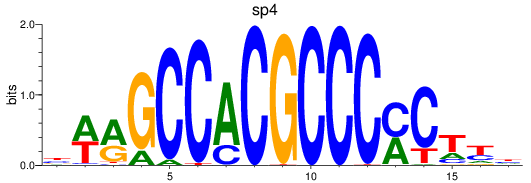
sp4
|
ENSDARG00000005186 | sp4 transcription factor |
|
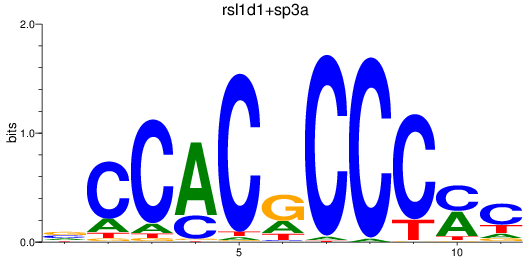
sp3a
|
ENSDARG00000001549 | sp3a transcription factor |
|
rsl1d1
|
ENSDARG00000005846 | Sp5 transcription factor b |
|
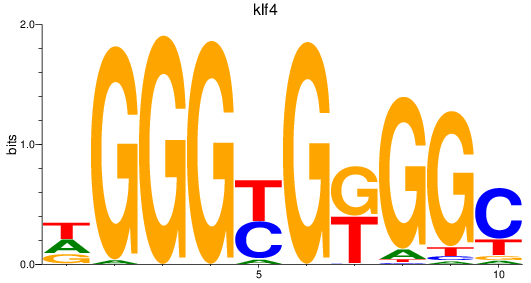
klf4
|
ENSDARG00000079922 | Kruppel-like factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp8b | dr11_v1_chr16_+_19536614_19536761 | -0.72 | 3.0e-16 | Click! |
| FO704755.1 | dr11_v1_chr6_+_3809346_3809346 | -0.40 | 6.0e-05 | Click! |
| sp8a | dr11_v1_chr19_-_2317558_2317558 | -0.37 | 2.6e-04 | Click! |
| sp3a | dr11_v1_chr9_+_2762270_2762415 | 0.33 | 9.9e-04 | Click! |
| klf4 | dr11_v1_chr21_-_435466_435466 | 0.18 | 8.1e-02 | Click! |
| sp4 | dr11_v1_chr19_+_2546775_2546775 | 0.17 | 9.4e-02 | Click! |
Activity profile of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4 motif
Sorted Z-values of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_17197546 | 64.69 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr12_+_46634736 | 39.91 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr10_-_41352502 | 35.37 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr20_-_54564018 | 32.84 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr3_+_1211242 | 31.75 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr24_-_11076400 | 30.86 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr21_-_32436679 | 29.10 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr10_-_1625080 | 29.06 |
ENSDART00000137285
|
nup155
|
nucleoporin 155 |
| chr16_-_17713859 | 28.51 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr22_+_11756040 | 27.41 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr21_-_11646878 | 27.13 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr25_-_22187397 | 26.61 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr21_-_43328056 | 26.56 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr5_-_1047504 | 26.44 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr22_+_661505 | 25.84 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr3_+_19299309 | 25.49 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr2_+_68789 | 25.17 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr15_+_17752928 | 24.50 |
ENSDART00000155314
|
si:ch211-213d14.2
|
si:ch211-213d14.2 |
| chr24_+_42132962 | 23.91 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr6_-_60104628 | 23.61 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr24_+_37449893 | 23.57 |
ENSDART00000164405
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr24_+_35387517 | 23.47 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr22_+_1006573 | 23.27 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr1_-_59232267 | 23.26 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr19_+_48111285 | 23.08 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr21_+_25236297 | 22.67 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr6_-_7776612 | 22.25 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr2_+_30379650 | 22.21 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr16_+_28754403 | 22.18 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr10_-_2942900 | 21.54 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr15_+_28410664 | 21.51 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr22_+_835728 | 21.37 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr8_+_23213320 | 21.10 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr21_-_39024754 | 20.72 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr2_+_35880600 | 20.56 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr15_+_46344655 | 20.48 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr6_+_102506 | 20.47 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr24_+_42127983 | 20.09 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr18_+_6558338 | 19.92 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr25_+_4635355 | 19.83 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr22_+_661711 | 19.65 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr17_+_27456804 | 18.43 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr25_+_37443194 | 18.40 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr22_+_883678 | 18.24 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr2_-_51109476 | 18.21 |
ENSDART00000168286
|
si:ch73-52e5.1
|
si:ch73-52e5.1 |
| chr19_+_9305964 | 17.88 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr16_+_53455638 | 17.65 |
ENSDART00000045792
ENSDART00000154189 |
rbm24b
|
RNA binding motif protein 24b |
| chr22_+_24645325 | 17.46 |
ENSDART00000159531
|
lpar3
|
lysophosphatidic acid receptor 3 |
| chr14_+_7048930 | 17.45 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr12_-_4331117 | 17.28 |
ENSDART00000008893
|
ca15a
|
carbonic anhydrase XVa |
| chr6_+_41503854 | 17.28 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr24_-_36593876 | 17.22 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr3_-_40276057 | 17.18 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr2_+_413370 | 17.16 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr11_+_13524504 | 16.97 |
ENSDART00000139889
|
arrdc2
|
arrestin domain containing 2 |
| chr12_-_4540564 | 16.94 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr18_-_6803424 | 16.90 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr7_+_1473929 | 16.86 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr23_-_10254288 | 16.82 |
ENSDART00000081215
|
krt8
|
keratin 8 |
| chr21_-_308852 | 16.67 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr22_+_11775269 | 16.64 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr10_-_4980150 | 16.43 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr16_-_12316979 | 15.87 |
ENSDART00000182392
|
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr9_-_34269066 | 15.60 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr12_+_23424108 | 15.54 |
ENSDART00000077732
|
bambia
|
BMP and activin membrane-bound inhibitor (Xenopus laevis) homolog a |
| chr1_-_53907092 | 15.46 |
ENSDART00000007732
|
capn9
|
calpain 9 |
| chr22_-_607812 | 15.44 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr24_-_29777413 | 15.42 |
ENSDART00000087724
|
wu:fj05g07
|
wu:fj05g07 |
| chr20_-_7293837 | 15.26 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr5_-_23696926 | 15.25 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr19_+_43119698 | 15.20 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr3_-_32541033 | 15.16 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr4_+_57093908 | 15.14 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr3_+_36972586 | 15.05 |
ENSDART00000102784
|
si:ch211-18i17.2
|
si:ch211-18i17.2 |
| chr20_-_9580446 | 14.92 |
ENSDART00000014168
|
zfp36l1b
|
zinc finger protein 36, C3H type-like 1b |
| chr24_+_17005647 | 14.87 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr3_+_32492467 | 14.81 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr9_+_1654284 | 14.79 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr15_+_46357080 | 14.68 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr4_-_4612116 | 14.59 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr25_+_3104959 | 14.45 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr5_-_1047222 | 14.41 |
ENSDART00000181112
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr8_+_47099033 | 14.30 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr22_+_396840 | 14.16 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr21_-_32060993 | 14.14 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr3_+_29469283 | 14.14 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr12_+_48841182 | 14.07 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr1_-_54107321 | 13.81 |
ENSDART00000148382
ENSDART00000150357 |
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr1_+_59538755 | 13.71 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr3_+_726000 | 13.56 |
ENSDART00000158510
|
dicp1.17
|
diverse immunoglobulin domain-containing protein 1.17 |
| chr24_-_32582378 | 13.49 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr25_+_34013093 | 13.48 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr4_+_78008298 | 13.41 |
ENSDART00000174237
|
cct2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr21_+_1647990 | 13.39 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr14_+_97017 | 13.38 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr8_+_6576940 | 13.32 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr19_-_6193448 | 13.30 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr22_+_20720808 | 13.25 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr19_+_19989380 | 13.00 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr25_+_186583 | 12.83 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr25_+_6122823 | 12.70 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr24_+_38671054 | 12.69 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr15_+_46356879 | 12.50 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr25_+_3306858 | 12.44 |
ENSDART00000137077
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr20_+_26538137 | 12.33 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr11_-_25384213 | 12.28 |
ENSDART00000103650
|
mafbb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
| chr12_+_48841419 | 12.25 |
ENSDART00000125331
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr15_-_20468302 | 12.11 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr1_-_59216197 | 12.07 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr8_-_53490376 | 12.04 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr18_+_6558146 | 12.03 |
ENSDART00000169401
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr7_-_52498175 | 12.00 |
ENSDART00000129769
|
cgnl1
|
cingulin-like 1 |
| chr8_+_53051701 | 11.91 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr18_+_44649804 | 11.89 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr2_+_55365727 | 11.89 |
ENSDART00000162943
|
FP245456.1
|
|
| chr7_+_38349667 | 11.88 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr20_+_34400715 | 11.86 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr8_+_8927870 | 11.80 |
ENSDART00000081985
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr1_-_51734524 | 11.77 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr17_+_51746830 | 11.70 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr21_+_43328685 | 11.68 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr23_-_10177442 | 11.65 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr23_-_5818992 | 11.62 |
ENSDART00000148730
|
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr22_-_16275236 | 11.51 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr6_-_39313027 | 11.40 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr3_+_5575313 | 11.27 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr17_+_389218 | 11.22 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr8_+_52637507 | 11.11 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr21_-_13856689 | 11.09 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr8_+_19356072 | 10.99 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr8_+_51958968 | 10.97 |
ENSDART00000139509
ENSDART00000186544 |
gna14a
|
guanine nucleotide binding protein (G protein), alpha 14a |
| chr19_-_11031145 | 10.96 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr4_-_17409533 | 10.94 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr13_-_42673978 | 10.87 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr16_+_23431189 | 10.81 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr6_+_3334710 | 10.81 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr14_+_4151379 | 10.81 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr5_+_46293850 | 10.62 |
ENSDART00000140636
|
si:ch211-130m23.2
|
si:ch211-130m23.2 |
| chr5_+_66132394 | 10.55 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr10_-_15048781 | 10.54 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr23_-_31436524 | 10.53 |
ENSDART00000140519
|
zgc:153284
|
zgc:153284 |
| chr6_+_49926115 | 10.50 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr23_-_2513300 | 10.49 |
ENSDART00000067652
|
snai1b
|
snail family zinc finger 1b |
| chr23_-_31645760 | 10.40 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr6_-_607063 | 10.37 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr18_-_48547564 | 10.34 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr3_+_32410746 | 10.19 |
ENSDART00000025496
|
rras
|
RAS related |
| chr21_-_5056812 | 10.15 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr14_+_26439227 | 10.12 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr10_+_31809226 | 10.12 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr10_-_44017642 | 9.97 |
ENSDART00000135240
ENSDART00000014669 |
acads
|
acyl-CoA dehydrogenase short chain |
| chr5_+_72152813 | 9.94 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr16_+_23282655 | 9.91 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr4_-_20181964 | 9.88 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr8_+_29962635 | 9.87 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr19_-_1855368 | 9.84 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr2_+_48073972 | 9.81 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr25_+_3306620 | 9.77 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr10_-_15053507 | 9.64 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr13_-_36582341 | 9.58 |
ENSDART00000137335
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr8_-_2616326 | 9.51 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr12_+_695619 | 9.46 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr25_-_37284370 | 9.44 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr19_+_19988869 | 9.37 |
ENSDART00000151024
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr2_-_48826707 | 9.32 |
ENSDART00000134711
|
svilb
|
supervillin b |
| chr23_-_45398622 | 9.32 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr25_+_10547228 | 9.31 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr2_+_48074243 | 9.27 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr25_+_34915762 | 9.25 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr11_+_11719575 | 9.20 |
ENSDART00000003891
|
jupa
|
junction plakoglobin a |
| chr5_-_68022631 | 9.18 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr17_-_20558961 | 9.15 |
ENSDART00000155993
|
sh3pxd2ab
|
SH3 and PX domains 2Ab |
| chr7_+_58730201 | 9.09 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr6_+_50451337 | 9.08 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr24_-_4450238 | 9.02 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr17_+_24615091 | 9.01 |
ENSDART00000064739
|
rpl13a
|
ribosomal protein L13a |
| chr15_+_36105581 | 9.00 |
ENSDART00000184099
|
col4a3
|
collagen, type IV, alpha 3 |
| chr21_-_11632403 | 9.00 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr8_-_4100365 | 8.95 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr8_+_7033049 | 8.93 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr9_+_30421489 | 8.88 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr19_-_6193067 | 8.87 |
ENSDART00000092656
ENSDART00000140347 |
erf
|
Ets2 repressor factor |
| chr2_-_51630555 | 8.85 |
ENSDART00000171746
|
pigr
|
polymeric immunoglobulin receptor |
| chr23_-_45407631 | 8.83 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr22_-_27136451 | 8.83 |
ENSDART00000153589
|
si:dkey-16m19.1
|
si:dkey-16m19.1 |
| chr7_-_10560964 | 8.73 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr5_-_46505691 | 8.62 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr13_+_25410594 | 8.61 |
ENSDART00000112092
|
calhm3
|
calcium homeostasis modulator 3 |
| chr19_+_46113828 | 8.61 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr21_+_15713097 | 8.61 |
ENSDART00000015841
|
gstt1b
|
glutathione S-transferase theta 1b |
| chr6_-_40657653 | 8.56 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr8_-_38201415 | 8.54 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr5_+_4533244 | 8.54 |
ENSDART00000158826
|
CABZ01058650.1
|
Danio rerio thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 (LOC561325), mRNA. |
| chr1_+_22654875 | 8.53 |
ENSDART00000019698
ENSDART00000161874 |
anxa5b
|
annexin A5b |
| chr11_+_329687 | 8.51 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr25_-_35960229 | 8.50 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr25_-_16724913 | 8.50 |
ENSDART00000157075
|
galnt8a.2
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 2 |
| chr13_+_22480496 | 8.48 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr1_-_54100988 | 8.39 |
ENSDART00000192662
|
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr10_-_320153 | 8.38 |
ENSDART00000161493
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 29.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 6.7 | 6.7 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 6.6 | 26.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 6.2 | 43.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 6.1 | 18.3 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 5.8 | 17.5 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 5.7 | 17.2 | GO:0006565 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 4.8 | 23.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 4.6 | 18.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 4.4 | 22.2 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 4.2 | 25.5 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 4.0 | 12.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 4.0 | 15.9 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 3.6 | 10.8 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 3.5 | 24.4 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 3.4 | 23.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 3.1 | 15.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 3.0 | 12.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 3.0 | 9.0 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 3.0 | 8.9 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 2.8 | 8.4 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 2.8 | 19.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 2.7 | 8.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 2.7 | 13.5 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 2.7 | 16.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 2.7 | 24.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 2.6 | 7.8 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 2.6 | 7.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 2.5 | 12.7 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 2.5 | 32.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 2.5 | 10.0 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 2.3 | 16.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 2.3 | 6.8 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 2.2 | 2.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 2.2 | 8.9 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 2.1 | 8.6 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 2.1 | 10.5 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 2.1 | 8.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 2.1 | 16.7 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 2.1 | 10.3 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 2.0 | 6.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 2.0 | 6.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 2.0 | 4.0 | GO:1903428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 1.9 | 7.8 | GO:0002934 | desmosome organization(GO:0002934) |
| 1.9 | 5.8 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 1.9 | 17.4 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 1.9 | 5.6 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 1.8 | 7.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 1.8 | 7.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 1.8 | 21.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 1.7 | 12.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 1.7 | 12.2 | GO:1902623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 1.7 | 12.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 1.7 | 1.7 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 1.7 | 5.0 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 1.7 | 6.7 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 1.7 | 3.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 1.6 | 4.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.6 | 6.4 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 1.6 | 4.7 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.5 | 10.7 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 1.5 | 14.8 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 1.5 | 7.4 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 1.5 | 10.2 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 1.4 | 15.9 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.4 | 23.1 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 1.4 | 51.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 1.4 | 11.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 1.3 | 18.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 1.3 | 5.2 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 1.3 | 30.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 1.3 | 5.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.3 | 2.6 | GO:1902745 | positive regulation of lamellipodium organization(GO:1902745) |
| 1.2 | 3.7 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 1.2 | 1.2 | GO:0010878 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) |
| 1.2 | 58.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 1.2 | 3.6 | GO:0048785 | hatching gland development(GO:0048785) |
| 1.2 | 4.7 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 1.2 | 4.7 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 1.2 | 3.5 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 1.2 | 6.9 | GO:0051818 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 1.1 | 11.4 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 1.1 | 11.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 1.1 | 4.5 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 1.1 | 7.8 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 1.1 | 8.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.1 | 23.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 1.0 | 21.0 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 1.0 | 4.2 | GO:0060437 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 1.0 | 3.1 | GO:0003071 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 1.0 | 15.4 | GO:0035904 | aorta development(GO:0035904) |
| 1.0 | 2.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 1.0 | 4.1 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 1.0 | 15.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 1.0 | 3.0 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.0 | 6.0 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.0 | 2.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.0 | 5.9 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 1.0 | 3.9 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 1.0 | 5.8 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 1.0 | 8.6 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.9 | 8.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.9 | 10.9 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.9 | 13.6 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.9 | 5.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.9 | 3.5 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.9 | 5.3 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.9 | 5.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.9 | 10.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.9 | 2.6 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.9 | 4.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.9 | 2.6 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.9 | 7.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.9 | 11.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.9 | 2.6 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.8 | 29.7 | GO:0007586 | digestion(GO:0007586) |
| 0.8 | 2.5 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.8 | 17.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.8 | 0.8 | GO:0061195 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.8 | 10.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.8 | 3.3 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.8 | 3.3 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.8 | 3.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.8 | 3.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.8 | 6.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.8 | 4.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.8 | 12.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.8 | 2.4 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.8 | 5.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.8 | 3.1 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.7 | 11.2 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.7 | 15.5 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.7 | 3.0 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.7 | 2.2 | GO:0072068 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.7 | 8.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.7 | 2.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.7 | 2.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.7 | 3.6 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.7 | 10.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.7 | 4.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 2.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.7 | 3.4 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.7 | 3.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.7 | 16.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.7 | 8.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.7 | 13.4 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.7 | 2.7 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.6 | 5.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 2.6 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.6 | 3.2 | GO:0019478 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.6 | 7.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.6 | 1.9 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.6 | 3.8 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.6 | 4.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.6 | 4.5 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.6 | 5.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.6 | 9.4 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.6 | 2.5 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.6 | 9.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.6 | 3.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.6 | 6.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.6 | 8.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.6 | 4.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.6 | 1.8 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.6 | 4.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.6 | 8.6 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) |
| 0.6 | 6.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.6 | 5.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.6 | 2.8 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.6 | 2.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.6 | 1.7 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.6 | 5.6 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.6 | 5.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.6 | 3.9 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.5 | 16.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.5 | 4.3 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.5 | 3.2 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 0.5 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.5 | 1.6 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.5 | 2.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.5 | 5.7 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.5 | 3.0 | GO:0035989 | tendon development(GO:0035989) |
| 0.5 | 46.8 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.5 | 3.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.5 | 2.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.5 | 3.9 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.5 | 3.9 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.5 | 12.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.5 | 1.0 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.5 | 5.3 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.5 | 10.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 2.9 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.5 | 33.4 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.5 | 2.3 | GO:2000765 | regulation of cytoplasmic translation(GO:2000765) |
| 0.5 | 3.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.5 | 5.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.5 | 2.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.5 | 1.8 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.5 | 1.8 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.4 | 8.1 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.4 | 4.0 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.4 | 8.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.4 | 6.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 1.3 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.4 | 20.0 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.4 | 15.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.4 | 1.3 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.4 | 2.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.4 | 6.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.4 | 5.5 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.4 | 2.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.4 | 5.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.4 | 2.9 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.4 | 2.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.4 | 3.7 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.4 | 4.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.4 | 2.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.4 | 23.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.4 | 2.0 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.4 | 12.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.4 | 2.0 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.4 | 7.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.4 | 5.4 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.4 | 12.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 10.8 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.4 | 6.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.4 | 1.9 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.4 | 1.1 | GO:0042539 | hypotonic response(GO:0006971) hypotonic salinity response(GO:0042539) |
| 0.4 | 0.4 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.4 | 6.0 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.4 | 3.0 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.4 | 4.0 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 2.2 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.4 | 19.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.4 | 1.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 9.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.4 | 3.9 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.4 | 4.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 23.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.4 | 9.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.3 | 3.5 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.3 | 1.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.3 | 3.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.3 | 1.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.3 | 4.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 4.1 | GO:0002757 | immune response-activating signal transduction(GO:0002757) |
| 0.3 | 3.1 | GO:0030323 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.3 | 12.5 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.3 | 2.7 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.3 | 3.0 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.3 | 3.6 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.3 | 1.6 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.3 | 2.6 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.3 | 4.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 19.1 | GO:0042493 | response to drug(GO:0042493) |
| 0.3 | 2.8 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.3 | 1.6 | GO:0050680 | negative regulation of endothelial cell proliferation(GO:0001937) negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.3 | 7.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.3 | 1.9 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.3 | 3.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.3 | 3.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 0.9 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.3 | 1.2 | GO:0051149 | positive regulation of muscle cell differentiation(GO:0051149) |
| 0.3 | 1.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.3 | 5.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 4.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.3 | 1.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 0.6 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.3 | 8.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.3 | 3.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.3 | 6.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 | 1.7 | GO:0060149 | positive regulation of translational initiation(GO:0045948) negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.3 | 19.7 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.3 | 1.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.3 | 18.7 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.3 | 0.8 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.3 | 2.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 16.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.3 | 2.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 1.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 9.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.3 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 6.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 1.7 | GO:1900094 | BMP signaling pathway involved in determination of left/right symmetry(GO:0003154) BMP signaling pathway involved in heart jogging(GO:0003303) negative regulation of endodermal cell fate specification(GO:0042664) regulation of transcription from RNA polymerase II promoter involved in determination of left/right symmetry(GO:1900094) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 31.2 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.2 | 55.5 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.2 | 1.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 1.4 | GO:0019859 | thymine catabolic process(GO:0006210) valine catabolic process(GO:0006574) thymine metabolic process(GO:0019859) |
| 0.2 | 1.3 | GO:0033146 | regulation of intracellular steroid hormone receptor signaling pathway(GO:0033143) regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.2 | 0.9 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 1.8 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 6.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 4.8 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.2 | 0.9 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 | 11.6 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.2 | 3.7 | GO:0007032 | endosome organization(GO:0007032) |
| 0.2 | 9.8 | GO:1990266 | granulocyte migration(GO:0097530) neutrophil migration(GO:1990266) |
| 0.2 | 2.1 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.2 | 0.6 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 6.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.7 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 3.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 1.2 | GO:0001840 | neural plate development(GO:0001840) |
| 0.2 | 5.6 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.2 | 0.4 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.2 | 9.5 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.2 | 1.5 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.2 | 1.1 | GO:0071347 | interleukin-1-mediated signaling pathway(GO:0070498) cellular response to interleukin-1(GO:0071347) |
| 0.2 | 3.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.2 | 7.4 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.2 | 9.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.2 | 3.6 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.2 | 5.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 1.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 1.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 0.9 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.2 | 1.6 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 0.5 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 1.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 2.6 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.2 | 0.5 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 2.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.2 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 1.5 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 6.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 0.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 0.2 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.2 | 1.8 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.2 | 2.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 3.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 2.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 4.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 0.8 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 3.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 2.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.8 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.1 | 0.7 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.9 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 14.7 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.1 | 5.6 | GO:0071391 | response to estrogen(GO:0043627) cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.6 | GO:0097535 | lymphoid progenitor cell differentiation(GO:0002320) lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 5.3 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.1 | 4.4 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.1 | 1.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.6 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 2.8 | GO:0032508 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.1 | 0.7 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 5.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.9 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 8.7 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.1 | 0.7 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 4.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 2.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.9 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 3.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 2.7 | GO:0038127 | ERBB signaling pathway(GO:0038127) |
| 0.1 | 1.4 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 0.7 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 1.0 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 2.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 3.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 6.3 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.1 | 3.1 | GO:0001878 | response to yeast(GO:0001878) response to fungus(GO:0009620) |
| 0.1 | 1.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.6 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.1 | 1.3 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 3.4 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 0.5 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 1.0 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 3.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 6.6 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 1.0 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.1 | 0.5 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism metabolic process(GO:0044033) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.8 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 0.9 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.1 | 0.8 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 2.5 | GO:0042098 | T cell proliferation(GO:0042098) regulation of T cell proliferation(GO:0042129) |
| 0.1 | 1.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 1.8 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 1.1 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 1.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 2.0 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 2.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.1 | 1.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 6.1 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 10.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.1 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 20.2 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.1 | 2.3 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 6.1 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.1 | 0.9 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.1 | 0.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 2.8 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 1.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 6.1 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.1 | 3.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 2.1 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 6.8 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 0.9 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 2.2 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.1 | 2.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 1.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 2.9 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.4 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 0.7 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.1 | 0.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 1.5 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.1 | 2.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.1 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 2.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.5 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 5.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.7 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.6 | GO:0001573 | ganglioside metabolic process(GO:0001573) ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.2 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 11.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 2.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 3.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 1.1 | GO:0002440 | production of molecular mediator of immune response(GO:0002440) |
| 0.0 | 4.8 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.4 | GO:0003159 | morphogenesis of an endothelium(GO:0003159) endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 20.1 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 23.2 | GO:0010941 | regulation of cell death(GO:0010941) |
| 0.0 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 2.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.2 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.0 | 0.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 2.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 1.0 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 1.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.7 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 2.0 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.6 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.8 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 1.3 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.0 | 0.3 | GO:0042159 | protein depalmitoylation(GO:0002084) lipoprotein catabolic process(GO:0042159) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0072028 | nephron morphogenesis(GO:0072028) |
| 0.0 | 0.8 | GO:0050817 | coagulation(GO:0050817) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 6.9 | GO:0043043 | translation(GO:0006412) peptide biosynthetic process(GO:0043043) |
| 0.0 | 1.0 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 6.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 3.3 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate metabolic process(GO:0030204) chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.3 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 29.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 4.5 | 49.7 | GO:0045095 | keratin filament(GO:0045095) |
| 3.5 | 14.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 3.5 | 17.7 | GO:0097433 | dense body(GO:0097433) |
| 3.2 | 12.8 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 3.0 | 56.8 | GO:0030057 | desmosome(GO:0030057) |
| 2.8 | 8.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 2.6 | 7.8 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 2.5 | 34.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 2.2 | 41.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 2.1 | 31.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 1.9 | 11.5 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 1.9 | 15.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.9 | 5.7 | GO:0098536 | deuterosome(GO:0098536) |
| 1.5 | 8.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 1.4 | 8.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 1.4 | 15.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 1.2 | 6.2 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 1.1 | 15.9 | GO:0042555 | MCM complex(GO:0042555) |
| 1.0 | 10.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.9 | 5.3 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.9 | 9.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.9 | 9.5 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.8 | 6.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.8 | 11.9 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.7 | 70.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.7 | 2.9 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.7 | 13.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.7 | 3.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.7 | 4.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.7 | 9.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.6 | 28.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.6 | 7.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 8.5 | GO:0070187 | telosome(GO:0070187) |
| 0.6 | 13.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.6 | 70.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.6 | 7.7 | GO:0002102 | podosome(GO:0002102) |
| 0.5 | 11.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.5 | 67.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.5 | 46.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.5 | 12.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.5 | 29.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.5 | 5.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.5 | 9.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 28.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.5 | 2.7 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.5 | 7.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.5 | 1.4 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 4.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 26.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 2.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 21.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 22.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.3 | 4.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.3 | 0.9 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 3.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 12.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.3 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 10.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 22.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.3 | 23.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.3 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 1.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 4.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 2.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 6.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 3.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 4.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 12.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 54.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 0.9 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.2 | 1.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 4.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 13.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.2 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 2.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 43.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 49.9 | GO:0000785 | chromatin(GO:0000785) |
| 0.2 | 17.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 38.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.2 | 7.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 1.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 7.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 2.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 10.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 46.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 10.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 6.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 5.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 0.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 5.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.9 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 2.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 5.9 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.3 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.1 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 2.2 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.8 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 14.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 8.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 4.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 5.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 10.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 104.0 | GO:0005829 | cytosol(GO:0005829) |
| 0.1 | 1.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 88.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 6.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 3.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 3.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 6.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 22.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 3.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 13.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 4.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.9 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 3.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.8 | 63.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 7.4 | 36.9 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 6.4 | 31.9 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 6.4 | 25.5 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 6.2 | 43.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 5.7 | 17.2 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 4.7 | 23.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 4.4 | 13.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 4.2 | 12.5 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 4.1 | 16.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 4.1 | 16.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 4.0 | 12.0 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 3.5 | 17.7 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 3.5 | 27.8 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 3.1 | 18.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 3.1 | 15.3 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 3.0 | 42.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 2.8 | 8.4 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 2.8 | 19.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 2.7 | 13.4 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 2.6 | 18.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 2.2 | 8.9 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 2.2 | 17.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 2.1 | 29.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 2.1 | 6.3 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 2.1 | 6.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 2.0 | 8.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 2.0 | 6.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 2.0 | 8.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 2.0 | 30.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 1.8 | 7.4 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 1.8 | 16.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 1.8 | 14.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 1.7 | 8.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.7 | 11.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.6 | 6.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.6 | 4.7 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 1.5 | 41.4 | GO:0042805 | actinin binding(GO:0042805) |
| 1.5 | 4.6 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.5 | 7.5 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 1.5 | 5.8 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.5 | 10.2 | GO:0070728 | leucine binding(GO:0070728) |
| 1.4 | 8.6 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 1.4 | 12.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 1.4 | 39.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 1.4 | 4.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 1.3 | 17.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.3 | 3.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 1.3 | 6.5 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 1.2 | 12.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.2 | 10.9 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 1.2 | 8.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 1.2 | 15.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 1.1 | 5.7 | GO:0051903 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 1.1 | 3.4 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 1.1 | 6.8 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 1.0 | 29.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 1.0 | 9.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.0 | 25.3 | GO:0005112 | Notch binding(GO:0005112) |
| 1.0 | 9.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 1.0 | 3.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.0 | 12.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.0 | 2.9 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 1.0 | 25.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 1.0 | 8.6 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 1.0 | 11.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.0 | 21.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.9 | 9.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.9 | 3.7 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.9 | 12.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.9 | 8.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.9 | 20.5 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.9 | 14.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.9 | 29.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.8 | 3.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.8 | 23.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.8 | 12.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.8 | 7.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.8 | 4.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 3.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.7 | 3.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.7 | 10.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 3.0 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.7 | 8.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.7 | 22.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.7 | 9.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.7 | 11.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.7 | 4.0 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.7 | 8.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.7 | 10.0 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.7 | 2.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.6 | 2.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.6 | 15.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.6 | 18.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.6 | 6.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.6 | 17.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.6 | 16.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.6 | 7.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.6 | 4.8 | GO:0036122 | BMP binding(GO:0036122) |
| 0.6 | 5.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.6 | 8.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.6 | 5.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.6 | 5.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.6 | 3.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 2.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.6 | 3.9 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.5 | 14.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.5 | 1.6 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.5 | 1.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.5 | 7.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 2.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.5 | 6.8 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.5 | 2.6 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.5 | 21.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.5 | 4.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 4.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 15.1 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.5 | 13.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.5 | 12.8 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.5 | 3.8 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.5 | 15.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.5 | 2.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 2.7 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.5 | 1.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 6.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 4.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 1.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.4 | 3.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.4 | 1.8 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.4 | 1.7 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.4 | 3.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 5.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.4 | 8.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 9.2 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.4 | 2.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.4 | 3.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.4 | 3.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.4 | 8.8 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.4 | 9.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.4 | 17.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 8.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.4 | 2.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.4 | 1.5 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.4 | 4.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.4 | 48.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 7.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.4 | 12.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.4 | 1.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.4 | 6.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 9.2 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.3 | 4.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 3.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 6.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 2.9 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.3 | 4.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 8.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 5.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 3.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.3 | 4.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 2.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.3 | 0.9 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.3 | 5.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 3.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 24.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.3 | 1.1 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.3 | 1.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.3 | 14.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 2.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 2.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 5.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.3 | 20.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 2.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.3 | 9.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 10.0 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 1.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 2.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 8.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 12.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.2 | 1.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 3.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 8.7 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.2 | 0.7 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 1.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 94.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.2 | 3.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 1.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 43.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.2 | 25.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 5.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.2 | 1.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 12.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 1.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 20.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.8 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.2 | 1.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 3.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 1.9 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 10.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.2 | 37.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 41.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 1.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.7 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.2 | 1.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 2.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 0.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 1.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 3.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 1.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 5.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 1.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 0.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.9 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 4.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 2.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.2 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 3.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 49.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.2 | 1.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 1.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 45.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 3.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 4.0 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 1.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 10.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 6.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 1.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.8 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 2.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 2.5 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.8 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 2.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 8.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 7.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 7.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.7 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 1.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 21.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 15.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.1 | 7.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 10.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 10.7 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.1 | 1.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 3.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 1.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.5 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.1 | 5.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 2.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.7 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.4 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 4.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 4.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.7 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.1 | 2.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 3.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 9.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 6.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 5.1 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 3.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 3.0 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.1 | 43.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 1.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 4.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 99.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 1.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 0.4 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 7.8 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 4.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.0 | 2.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.8 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 9.9 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 7.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 7.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 1.7 | 75.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 1.7 | 20.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.7 | 105.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 1.1 | 27.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.9 | 7.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.8 | 13.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.8 | 6.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.7 | 9.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.7 | 8.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.6 | 36.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.6 | 43.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.6 | 5.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.5 | 17.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.5 | 42.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.5 | 18.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.5 | 1.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.5 | 5.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.4 | 2.2 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.4 | 12.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.4 | 16.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.4 | 9.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.4 | 14.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.4 | 3.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 3.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 15.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.3 | 5.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.3 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 2.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 10.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.3 | 4.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.3 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.3 | 5.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 3.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 1.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 3.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 1.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.2 | 4.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 1.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.2 | 3.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 6.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 2.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 3.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 2.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 1.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 5.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 14.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 5.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 18.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 3.2 | 41.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 3.0 | 62.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 2.8 | 25.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 1.8 | 31.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 1.6 | 9.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 1.4 | 20.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 1.3 | 5.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 1.2 | 23.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 1.2 | 3.7 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 1.2 | 3.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 1.1 | 15.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 1.1 | 10.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.1 | 8.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 1.0 | 15.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 1.0 | 1.0 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 1.0 | 17.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.9 | 22.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.8 | 30.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.7 | 5.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.7 | 6.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.7 | 7.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.7 | 15.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.7 | 49.6 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.7 | 10.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.6 | 12.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.6 | 8.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.6 | 7.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 6.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.6 | 11.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.5 | 4.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.5 | 13.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.5 | 29.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.5 | 15.8 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.5 | 10.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.5 | 3.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.5 | 10.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.5 | 5.0 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.4 | 18.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.4 | 3.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 15.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.4 | 3.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 9.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.4 | 25.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.4 | 3.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.3 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 1.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 34.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 8.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 4.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 4.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 6.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 5.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.2 | 4.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 3.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.2 | 4.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.2 | 4.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 7.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 7.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 5.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.2 | 2.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 4.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 4.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 4.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 4.8 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 0.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 21.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.2 | 15.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.2 | 1.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 2.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 9.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 6.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.5 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.1 | 1.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 9.4 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.1 | 8.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 2.9 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.3 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.7 | REACTOME TRANSCRIPTION COUPLED NER TC NER | Genes involved in Transcription-coupled NER (TC-NER) |
| 0.1 | 1.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.6 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.1 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 3.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.7 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |