Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
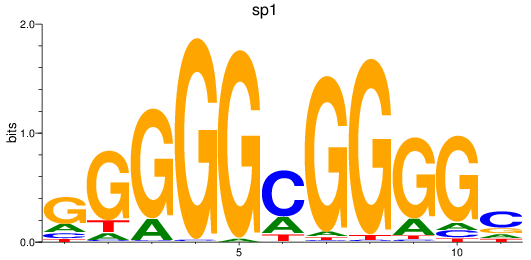
Results for sp1
Z-value: 2.07
Transcription factors associated with sp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp1
|
ENSDARG00000088347 | sp1 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp1 | dr11_v1_chr11_-_28747_28747 | 0.16 | 1.4e-01 | Click! |
Activity profile of sp1 motif
Sorted Z-values of sp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_5254699 | 29.85 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr13_+_1100197 | 23.87 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr8_-_4618653 | 23.70 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr7_+_568819 | 22.91 |
ENSDART00000173716
|
nrxn2b
|
neurexin 2b |
| chr2_+_58221163 | 22.80 |
ENSDART00000157939
|
FO704813.1
|
|
| chr21_+_26697536 | 22.12 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr7_+_73630751 | 20.88 |
ENSDART00000159745
|
PCP4L1
|
si:dkey-46i9.1 |
| chr15_-_47956388 | 20.20 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr3_-_62380146 | 19.79 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr11_-_32723851 | 18.65 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr7_+_7630409 | 18.11 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr16_-_44399335 | 16.01 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr8_+_7359294 | 15.42 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr10_-_31782616 | 15.16 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr22_+_12366516 | 14.84 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr11_+_25112269 | 14.80 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr22_+_5574952 | 14.79 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr2_-_44282796 | 14.57 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr11_+_25064519 | 14.53 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr1_-_14233815 | 14.35 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr1_-_10647484 | 14.12 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr9_-_44295071 | 13.99 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr1_-_18803919 | 13.72 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr3_+_24361096 | 13.58 |
ENSDART00000132387
|
pvalb6
|
parvalbumin 6 |
| chr1_-_26782573 | 13.45 |
ENSDART00000090611
|
sh3gl2a
|
SH3 domain containing GRB2 like 2a, endophilin A1 |
| chr1_-_59176949 | 13.44 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr1_-_10647307 | 13.34 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr17_+_15433518 | 13.21 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_-_43466317 | 13.10 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr17_+_15433671 | 12.97 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr4_-_5764255 | 12.96 |
ENSDART00000113864
|
faxca
|
failed axon connections homolog a |
| chr19_+_33701558 | 12.94 |
ENSDART00000147226
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr2_+_24177006 | 12.86 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr1_-_39943596 | 12.81 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr19_+_30633453 | 12.69 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr9_-_296169 | 12.51 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr11_+_25111846 | 12.40 |
ENSDART00000128705
ENSDART00000190058 |
ndrg3a
|
ndrg family member 3a |
| chr19_-_7450796 | 12.32 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr1_-_12278522 | 12.25 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr2_-_22535 | 12.24 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr10_+_15777258 | 12.19 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr12_-_25916530 | 12.13 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr11_-_103136 | 12.07 |
ENSDART00000173308
ENSDART00000162982 |
elmo2
|
engulfment and cell motility 2 |
| chr10_+_15777064 | 11.98 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr1_+_10720294 | 11.96 |
ENSDART00000139387
|
atp1b1b
|
ATPase Na+/K+ transporting subunit beta 1b |
| chr23_+_44732863 | 11.96 |
ENSDART00000160044
ENSDART00000172268 |
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr18_-_38087875 | 11.91 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr13_+_421231 | 11.87 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr25_-_13363286 | 11.86 |
ENSDART00000163735
ENSDART00000169119 |
ndrg4
|
NDRG family member 4 |
| chr16_+_10318893 | 11.82 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr2_-_31936966 | 11.82 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr13_+_4405282 | 11.74 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr8_-_30979494 | 11.69 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr13_+_12045758 | 11.58 |
ENSDART00000079398
ENSDART00000165467 ENSDART00000165880 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr22_-_11493236 | 11.55 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr2_+_24868010 | 11.48 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr5_-_13086616 | 11.46 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr25_+_21324588 | 11.45 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr2_-_9646857 | 11.40 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr6_+_34512313 | 11.31 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr17_-_20711735 | 11.29 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr9_+_35860975 | 11.25 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr20_+_33294428 | 11.24 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr8_-_14052349 | 11.20 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr16_+_7626535 | 11.16 |
ENSDART00000182670
ENSDART00000065514 ENSDART00000150212 |
stx12l
|
syntaxin 12, like |
| chr14_-_30642819 | 11.09 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr8_+_23165749 | 11.06 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr1_-_44704261 | 10.99 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr7_+_529522 | 10.69 |
ENSDART00000190811
|
nrxn2b
|
neurexin 2b |
| chr10_+_20128267 | 10.49 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr7_-_52876683 | 10.36 |
ENSDART00000172910
|
map1aa
|
microtubule-associated protein 1Aa |
| chr23_+_45579497 | 10.22 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr13_+_13693722 | 10.19 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr19_+_29798064 | 10.16 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr16_-_44349845 | 10.14 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr13_+_36764715 | 10.13 |
ENSDART00000111832
ENSDART00000085230 |
atl1
|
atlastin GTPase 1 |
| chr19_+_31771270 | 10.12 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr17_+_15535501 | 10.05 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr1_+_54908895 | 9.99 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr10_+_22012389 | 9.98 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr11_-_97817 | 9.92 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr9_+_22677503 | 9.91 |
ENSDART00000131429
ENSDART00000080005 ENSDART00000101756 ENSDART00000138148 |
itgb5
|
integrin, beta 5 |
| chr10_+_32561317 | 9.87 |
ENSDART00000109029
|
map6a
|
microtubule-associated protein 6a |
| chr20_-_19422496 | 9.86 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr1_-_14234076 | 9.86 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr8_-_25011157 | 9.73 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr6_+_32326074 | 9.71 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr3_+_22578369 | 9.70 |
ENSDART00000187695
ENSDART00000182678 ENSDART00000112270 |
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr20_-_34868814 | 9.67 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr21_-_43079161 | 9.66 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr20_-_39219537 | 9.64 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr13_+_11439486 | 9.64 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr18_-_38088099 | 9.64 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr20_-_31905968 | 9.57 |
ENSDART00000142806
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr12_-_37449396 | 9.56 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr6_-_13408680 | 9.55 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr23_-_4775446 | 9.49 |
ENSDART00000067531
|
syn2a
|
synapsin IIa |
| chr3_-_15264698 | 9.47 |
ENSDART00000111948
ENSDART00000142594 |
sez6l2
|
seizure related 6 homolog (mouse)-like 2 |
| chr5_+_64739762 | 9.44 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr16_-_52733384 | 9.43 |
ENSDART00000147236
ENSDART00000056101 |
azin1a
|
antizyme inhibitor 1a |
| chr10_-_32851847 | 9.43 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr12_-_14211293 | 9.39 |
ENSDART00000158399
|
avl9
|
AVL9 homolog (S. cerevisiase) |
| chr1_-_31505144 | 9.38 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr4_-_23839789 | 9.33 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr15_+_29393519 | 9.32 |
ENSDART00000193488
ENSDART00000112375 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr22_+_465269 | 9.29 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr14_-_26177156 | 9.29 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr7_+_50053339 | 9.24 |
ENSDART00000174308
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr15_+_1397811 | 9.16 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr10_-_35542071 | 9.07 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr13_-_43599898 | 8.94 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr16_-_42390441 | 8.92 |
ENSDART00000148475
|
cspg5a
|
chondroitin sulfate proteoglycan 5a |
| chr19_-_31802296 | 8.90 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr20_-_30035326 | 8.88 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr4_+_19534833 | 8.86 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr6_-_37622576 | 8.86 |
ENSDART00000154363
|
chst7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| chr15_-_33925851 | 8.81 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr13_-_22907260 | 8.79 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr11_-_36020005 | 8.78 |
ENSDART00000031993
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr14_+_5117072 | 8.72 |
ENSDART00000189628
|
nanos1
|
nanos homolog 1 |
| chr3_-_21118969 | 8.70 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr12_+_42436328 | 8.67 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr13_-_24218795 | 8.66 |
ENSDART00000136217
|
galnt2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 |
| chr24_+_31277360 | 8.59 |
ENSDART00000165993
|
f3a
|
coagulation factor IIIa |
| chr22_+_12361489 | 8.58 |
ENSDART00000182483
|
r3hdm1
|
R3H domain containing 1 |
| chr19_+_1097393 | 8.54 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr2_+_24177190 | 8.53 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr2_-_11912347 | 8.50 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr12_-_4651988 | 8.50 |
ENSDART00000182836
|
si:ch211-255p10.4
|
si:ch211-255p10.4 |
| chr20_-_53366137 | 8.48 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr4_-_890220 | 8.42 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr19_+_42716542 | 8.42 |
ENSDART00000144557
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr3_-_5964557 | 8.41 |
ENSDART00000184738
|
BX284638.1
|
|
| chr6_-_31348999 | 8.39 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr7_+_38762043 | 8.39 |
ENSDART00000036461
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr17_-_33289304 | 8.37 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr15_-_44601331 | 8.28 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr5_-_38506981 | 8.24 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr2_+_26656283 | 8.24 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr12_+_31729075 | 8.22 |
ENSDART00000152973
|
RNF157
|
si:dkey-49c17.3 |
| chr5_-_46273938 | 8.19 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr16_-_6821927 | 8.19 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr3_+_32425202 | 8.17 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr10_-_27046639 | 8.13 |
ENSDART00000041841
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr9_-_54840124 | 8.12 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr9_-_10145795 | 8.10 |
ENSDART00000004745
ENSDART00000143295 |
hnmt
|
histamine N-methyltransferase |
| chr10_-_1718395 | 8.09 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr20_+_37844035 | 7.97 |
ENSDART00000041397
|
flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr16_-_42390640 | 7.92 |
ENSDART00000193214
ENSDART00000102305 |
cspg5a
|
chondroitin sulfate proteoglycan 5a |
| chr6_-_14139503 | 7.85 |
ENSDART00000089577
|
cacnb4b
|
calcium channel, voltage-dependent, beta 4b subunit |
| chr2_+_25658112 | 7.80 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr14_+_1111332 | 7.79 |
ENSDART00000162660
|
si:ch1073-303k11.2
|
si:ch1073-303k11.2 |
| chr25_+_35375848 | 7.79 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr7_+_48460239 | 7.76 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr24_-_20599781 | 7.75 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr24_+_39158283 | 7.61 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr3_+_51684963 | 7.60 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr14_-_52552875 | 7.57 |
ENSDART00000170107
|
ppp2r2ba
|
protein phosphatase 2, regulatory subunit B, beta a |
| chr23_+_6232895 | 7.56 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr2_+_25657958 | 7.55 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr6_-_35738836 | 7.54 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr3_-_36440705 | 7.53 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr2_-_52550135 | 7.50 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr1_-_55873178 | 7.49 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr2_+_24867534 | 7.48 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr5_+_13427826 | 7.47 |
ENSDART00000083359
|
sec14l8
|
SEC14-like lipid binding 8 |
| chr3_-_6767440 | 7.46 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr10_-_36592486 | 7.44 |
ENSDART00000131693
|
slc1a3b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3b |
| chr7_+_38717624 | 7.41 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr22_-_20309283 | 7.37 |
ENSDART00000182125
ENSDART00000048775 |
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr2_+_26303627 | 7.32 |
ENSDART00000040278
|
efna2a
|
ephrin-A2a |
| chr12_-_35505610 | 7.31 |
ENSDART00000105518
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr12_+_9761685 | 7.30 |
ENSDART00000189522
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr14_+_33458294 | 7.27 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr15_+_40188076 | 7.20 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr23_+_2825940 | 7.18 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr18_-_50766660 | 7.12 |
ENSDART00000170663
ENSDART00000168601 |
zgc:158464
|
zgc:158464 |
| chr10_-_15405564 | 7.08 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr8_+_20157798 | 7.05 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr24_-_18467064 | 7.05 |
ENSDART00000081946
|
zgc:112332
|
zgc:112332 |
| chr5_+_38263240 | 7.04 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr16_+_32559821 | 7.03 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr12_+_32159272 | 7.03 |
ENSDART00000153167
|
hlfb
|
hepatic leukemia factor b |
| chr14_+_31865099 | 6.96 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr6_+_59967994 | 6.96 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr9_+_6082793 | 6.93 |
ENSDART00000192045
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr4_-_789645 | 6.92 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr17_-_31058900 | 6.91 |
ENSDART00000134998
ENSDART00000104307 ENSDART00000172721 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr16_-_45058919 | 6.90 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr18_+_36625490 | 6.89 |
ENSDART00000148032
|
clasrp
|
CLK4-associating serine/arginine rich protein |
| chr8_+_16025554 | 6.86 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr14_+_22172047 | 6.84 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr2_-_18830722 | 6.77 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr2_-_39759059 | 6.77 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr6_-_58828398 | 6.69 |
ENSDART00000090634
|
kif5ab
|
kinesin family member 5A, b |
| chr1_-_38709551 | 6.68 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr6_-_15604417 | 6.65 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr6_-_15604157 | 6.59 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr15_+_5901970 | 6.54 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr3_+_46459540 | 6.53 |
ENSDART00000188150
|
si:ch211-66e2.5
|
si:ch211-66e2.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 19.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 6.0 | 29.8 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 3.7 | 14.8 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 3.6 | 25.1 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 3.4 | 20.6 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 3.4 | 10.1 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 2.9 | 11.6 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 2.9 | 8.6 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 2.8 | 14.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 2.7 | 8.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 2.5 | 15.2 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 2.5 | 7.5 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 2.5 | 12.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 2.5 | 54.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 2.4 | 7.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 2.4 | 7.1 | GO:1903332 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 2.3 | 6.8 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 2.2 | 11.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 2.1 | 6.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 2.1 | 12.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 2.0 | 8.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 2.0 | 8.0 | GO:0097037 | heme export(GO:0097037) |
| 1.9 | 5.6 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 1.8 | 9.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 1.8 | 5.4 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 1.7 | 14.0 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 1.7 | 8.5 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 1.7 | 32.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 1.7 | 8.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 1.6 | 4.9 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 1.6 | 9.6 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 1.6 | 4.8 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 1.6 | 15.8 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 1.5 | 4.6 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 1.5 | 15.4 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 1.5 | 4.6 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.5 | 6.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 1.5 | 4.5 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 1.5 | 14.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 1.4 | 5.7 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 1.4 | 4.2 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 1.4 | 5.6 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 1.3 | 13.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.3 | 5.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 1.3 | 3.9 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 1.3 | 5.2 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 1.3 | 32.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 1.3 | 3.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.3 | 7.5 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 1.2 | 6.0 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.2 | 5.9 | GO:0034505 | tooth mineralization(GO:0034505) |
| 1.2 | 3.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 1.2 | 8.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 1.1 | 11.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 1.1 | 3.4 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 1.1 | 3.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 1.1 | 5.4 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 1.1 | 3.2 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 1.0 | 6.3 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 1.0 | 9.4 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 1.0 | 5.2 | GO:0045056 | transcytosis(GO:0045056) |
| 1.0 | 3.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 1.0 | 6.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 1.0 | 5.8 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 1.0 | 2.9 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.9 | 7.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.9 | 4.5 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.9 | 8.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.9 | 14.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.9 | 2.6 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.8 | 4.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.8 | 11.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.8 | 14.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.8 | 38.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.8 | 7.8 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.8 | 6.9 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.8 | 7.5 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.7 | 2.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.7 | 3.0 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.7 | 8.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.7 | 5.8 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.7 | 6.3 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.7 | 25.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.7 | 3.4 | GO:0040033 | miRNA metabolic process(GO:0010586) miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.7 | 8.7 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.7 | 3.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.7 | 2.6 | GO:0015677 | copper ion import(GO:0015677) |
| 0.6 | 4.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.6 | 1.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.6 | 8.8 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.6 | 5.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.6 | 6.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.6 | 3.7 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.6 | 3.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.6 | 3.7 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.6 | 7.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.6 | 2.4 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.6 | 4.2 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.6 | 4.8 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.6 | 3.5 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.6 | 1.7 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.6 | 3.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.6 | 1.7 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.6 | 2.3 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.6 | 21.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.6 | 2.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.6 | 3.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.6 | 2.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.6 | 11.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 2.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.5 | 2.7 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.5 | 9.6 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.5 | 1.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.5 | 3.7 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.5 | 15.4 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.5 | 3.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.5 | 5.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.5 | 6.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.5 | 10.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.5 | 5.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.5 | 8.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.5 | 4.6 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.5 | 15.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.5 | 2.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.5 | 5.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.5 | 4.5 | GO:0033273 | response to vitamin(GO:0033273) |
| 0.5 | 3.0 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.5 | 9.4 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.5 | 2.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.5 | 2.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.5 | 3.9 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.5 | 4.8 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.5 | 1.9 | GO:0019075 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.5 | 14.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.5 | 2.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.5 | 7.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.5 | 0.9 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.4 | 0.9 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.4 | 1.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.4 | 4.9 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 4.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.4 | 2.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 1.3 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.4 | 2.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.4 | 8.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.4 | 7.9 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.4 | 10.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.4 | 2.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.4 | 3.6 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.4 | 8.4 | GO:0007020 | microtubule nucleation(GO:0007020) exit from mitosis(GO:0010458) microtubule anchoring(GO:0034453) |
| 0.4 | 2.4 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.4 | 4.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.4 | 7.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.4 | 5.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 2.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.4 | 0.8 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.4 | 2.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.4 | 2.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.4 | 4.1 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 14.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.4 | 1.8 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.4 | 2.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.4 | 1.4 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.3 | 6.6 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.3 | 12.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.3 | 5.8 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 2.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.3 | 4.4 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.3 | 0.7 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.3 | 1.3 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.3 | 1.0 | GO:0070640 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D metabolic process(GO:0042359) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) vitamin D3 metabolic process(GO:0070640) |
| 0.3 | 5.2 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.3 | 4.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 1.0 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.3 | 4.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.3 | 4.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.3 | 4.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.3 | 5.8 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.3 | 2.2 | GO:0036268 | swimming(GO:0036268) |
| 0.3 | 4.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 2.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 7.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.3 | 2.5 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.3 | 0.9 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.3 | 1.8 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.3 | 1.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.3 | 0.9 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.3 | 11.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.3 | 5.1 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.3 | 5.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.3 | 1.4 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.3 | 28.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.3 | 8.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.3 | 5.0 | GO:1903321 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.3 | 1.9 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.3 | 9.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 2.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.3 | 3.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.3 | 2.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 0.8 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 2.9 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.3 | 5.0 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.3 | 4.2 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.3 | 7.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.3 | 0.8 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 1.3 | GO:0046456 | icosanoid biosynthetic process(GO:0046456) fatty acid derivative biosynthetic process(GO:1901570) |
| 0.3 | 6.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.2 | 1.7 | GO:0043931 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.2 | 3.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 3.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 10.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.2 | 2.9 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 0.7 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.2 | 7.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.2 | 2.4 | GO:0042044 | fluid transport(GO:0042044) |
| 0.2 | 4.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 5.3 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.2 | 3.9 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.2 | 2.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.2 | 1.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 | 2.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.2 | 7.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.7 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 5.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 19.4 | GO:0042552 | myelination(GO:0042552) |
| 0.2 | 5.9 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.2 | 15.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.2 | 10.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.2 | 1.3 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.2 | 2.9 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.2 | 9.0 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.2 | 1.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 4.9 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.2 | 4.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 3.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.2 | 8.4 | GO:0003401 | axis elongation(GO:0003401) |
| 0.2 | 2.2 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.2 | 5.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 1.4 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 3.1 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.2 | 1.6 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 2.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 16.0 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.2 | 9.5 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.2 | 3.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.2 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.9 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 2.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 1.8 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.2 | 5.6 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 2.8 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 4.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 0.3 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.2 | 8.5 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.2 | 2.8 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 1.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 2.2 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.2 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 0.9 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.2 | 1.8 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 10.6 | GO:0003407 | neural retina development(GO:0003407) |
| 0.2 | 0.6 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 1.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 3.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 1.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 1.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.6 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 31.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 14.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 4.8 | GO:0001764 | neuron migration(GO:0001764) |
| 0.1 | 1.7 | GO:0050890 | cognition(GO:0050890) |
| 0.1 | 16.2 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.1 | 1.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 1.3 | GO:0010138 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.1 | 2.9 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 1.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 1.3 | GO:0006188 | IMP biosynthetic process(GO:0006188) |
| 0.1 | 2.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.4 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 0.5 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 7.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 2.9 | GO:0015833 | peptide transport(GO:0015833) |
| 0.1 | 12.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 4.3 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.1 | 6.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 5.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 12.8 | GO:0060026 | convergent extension(GO:0060026) |
| 0.1 | 37.4 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.1 | 5.6 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.1 | 7.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 1.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 2.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.7 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 9.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 0.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 2.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 4.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.4 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.1 | 3.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.7 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.3 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 2.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.3 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 0.4 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 3.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 3.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.1 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.1 | 11.2 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 10.0 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 5.6 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 6.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.6 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.3 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 3.6 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.1 | 0.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 1.6 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 0.5 | GO:1902742 | apoptotic process involved in morphogenesis(GO:0060561) apoptotic process involved in development(GO:1902742) |
| 0.1 | 2.3 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 1.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.4 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 18.7 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.1 | 1.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 3.7 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.1 | 0.6 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.1 | 0.1 | GO:2000252 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 10.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 0.5 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.1 | 2.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.5 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 11.1 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.1 | 1.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 1.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.5 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 5.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 3.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 4.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.9 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.7 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 14.7 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.3 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 10.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 3.0 | GO:0030509 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.3 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 1.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 1.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.6 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.5 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 2.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.6 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 6.4 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 1.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.5 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 22.2 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.0 | 0.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.5 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 23.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 3.9 | 39.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 2.4 | 41.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 2.4 | 7.1 | GO:0072380 | TRC complex(GO:0072380) |
| 2.2 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 1.9 | 7.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.9 | 11.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 1.8 | 51.6 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 1.6 | 29.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 1.5 | 7.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 1.3 | 9.4 | GO:0016234 | inclusion body(GO:0016234) |
| 1.2 | 29.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 1.0 | 7.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.0 | 12.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 1.0 | 4.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.9 | 3.7 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.9 | 4.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.9 | 4.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.8 | 9.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.8 | 5.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.8 | 3.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.7 | 2.8 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.7 | 2.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.7 | 10.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.7 | 4.6 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.6 | 2.4 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.6 | 22.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.6 | 16.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.6 | 2.3 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.6 | 87.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.5 | 9.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.5 | 13.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.5 | 7.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.5 | 9.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.5 | 19.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.5 | 3.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.5 | 19.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.5 | 15.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.5 | 6.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 4.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 6.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.4 | 5.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.4 | 35.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 5.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.4 | 36.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.4 | 28.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.4 | 11.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 2.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 12.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.3 | 4.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 20.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 2.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 3.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 6.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 0.9 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.3 | 1.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 5.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 2.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 1.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.3 | 12.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 4.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 6.7 | GO:0060293 | germ plasm(GO:0060293) |
| 0.2 | 13.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 2.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 5.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 2.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.7 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.2 | 5.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 11.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 3.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.8 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 0.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 139.0 | GO:0043005 | neuron projection(GO:0043005) |
| 0.2 | 2.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 0.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 2.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 9.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 12.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 3.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 3.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 2.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 14.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 1.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 3.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.2 | 9.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 1.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 5.7 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 2.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 5.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.6 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 1.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.4 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 2.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 2.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 2.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 2.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 5.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 10.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 5.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 11.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 18.9 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 1.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 3.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 6.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 6.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.5 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 3.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 1.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 2.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 3.3 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 5.6 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 3.4 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 11.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.5 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.2 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 2.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.1 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 3.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 77.2 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.6 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 3.9 | 11.6 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 3.5 | 14.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 3.5 | 6.9 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 3.2 | 9.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 3.0 | 9.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 2.8 | 8.5 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 2.8 | 17.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.5 | 15.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 2.5 | 5.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 2.3 | 6.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 2.2 | 8.9 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 2.1 | 32.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 2.0 | 41.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.9 | 7.7 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 1.9 | 9.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 1.9 | 13.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.6 | 32.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 1.5 | 4.6 | GO:0046978 | TAP1 binding(GO:0046978) |
| 1.5 | 17.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 1.4 | 34.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 1.4 | 4.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 1.4 | 5.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 1.1 | 4.6 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 1.1 | 4.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 1.0 | 59.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 1.0 | 3.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.0 | 8.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.0 | 22.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.9 | 7.6 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.9 | 5.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.9 | 5.6 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.9 | 2.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.9 | 3.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.9 | 29.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.9 | 21.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.9 | 6.9 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.8 | 16.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.8 | 10.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.8 | 2.5 | GO:0031835 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.8 | 2.4 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.8 | 6.5 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.8 | 9.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.8 | 38.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.8 | 4.5 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.7 | 3.7 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.7 | 3.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.7 | 7.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.7 | 2.0 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.7 | 2.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.7 | 5.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.7 | 2.6 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.7 | 7.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.6 | 10.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.6 | 1.9 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.6 | 13.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.6 | 2.5 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.6 | 2.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.6 | 7.9 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.6 | 5.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.6 | 41.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.6 | 5.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.6 | 1.7 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.6 | 9.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.6 | 5.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.6 | 8.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 11.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 3.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.5 | 4.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 4.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.5 | 2.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.5 | 4.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.5 | 10.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.5 | 2.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 6.1 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.5 | 17.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 8.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.5 | 3.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.5 | 2.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.5 | 13.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.5 | 7.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.5 | 4.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.5 | 13.4 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.5 | 12.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 3.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 4.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 3.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.4 | 5.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.4 | 10.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.4 | 11.4 | GO:0010857 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.4 | 1.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.4 | 7.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.4 | 2.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.4 | 3.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 20.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.4 | 4.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.4 | 12.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.4 | 2.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 22.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.4 | 2.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.4 | 3.0 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.4 | 49.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.4 | 3.7 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.4 | 2.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.4 | 12.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 4.5 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.3 | 3.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 1.3 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.3 | 3.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 2.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.3 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.3 | 1.0 | GO:0031073 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.3 | 1.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.3 | 5.5 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.3 | 15.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.3 | 2.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 3.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 2.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.3 | 7.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.3 | 4.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 3.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 4.7 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.3 | 1.7 | GO:0030331 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.3 | 3.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 1.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 5.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.3 | 4.9 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.3 | 7.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.3 | 4.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 2.9 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 1.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.3 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 5.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 11.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 3.0 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 3.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 6.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 2.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 1.8 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.2 | 3.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 3.9 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.2 | 6.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 9.4 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.2 | 2.9 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.2 | 6.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 0.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 11.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 1.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 2.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 5.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 1.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 9.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 2.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 2.3 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.2 | 4.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 9.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 4.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 0.5 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 1.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 1.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 0.7 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.2 | 1.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 4.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.2 | 0.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 0.9 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 51.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.2 | 22.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 55.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 5.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 6.7 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 10.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.6 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 6.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 15.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 6.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 2.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 3.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.7 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 10.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 3.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 1.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 9.5 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 9.2 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 30.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 2.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 25.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 2.2 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 3.8 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 13.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 62.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 11.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 2.5 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 14.9 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 6.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.4 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.1 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 5.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.9 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 1.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.5 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0017069 | snRNA binding(GO:0017069) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 20.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 1.1 | 5.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.9 | 18.8 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.9 | 9.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.8 | 16.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.7 | 5.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.6 | 9.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.6 | 20.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.6 | 19.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.5 | 19.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.5 | 3.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 6.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.4 | 10.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.4 | 9.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 6.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 3.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 5.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 6.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 9.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 9.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 5.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 2.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 4.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 5.7 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 3.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 3.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 1.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 2.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 3.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 2.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 1.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 4.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.2 | 4.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 5.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 2.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 5.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 6.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 48.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 1.8 | 9.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 1.8 | 7.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 1.8 | 29.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 1.4 | 16.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 1.3 | 10.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 1.2 | 15.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 1.0 | 4.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.9 | 8.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.8 | 7.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.7 | 4.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.7 | 4.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.6 | 3.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.6 | 10.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 4.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.5 | 6.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 5.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 6.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.4 | 6.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.4 | 8.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.4 | 4.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.4 | 3.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.4 | 5.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 8.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.4 | 3.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 7.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.4 | 10.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 3.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 4.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 1.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 2.9 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.3 | 2.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.3 | 6.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.3 | 3.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.3 | 4.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.3 | 2.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.3 | 2.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.3 | 1.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.2 | 2.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 4.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 13.5 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.2 | 2.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 8.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 6.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 3.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 1.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 5.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 2.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 9.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 4.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 14.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 2.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 9.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 5.3 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 2.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 3.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |