Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
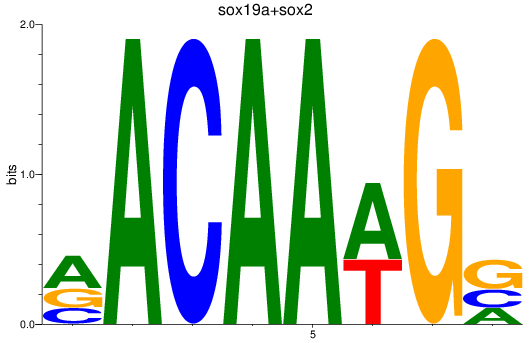
Results for sox19a+sox2
Z-value: 3.27
Transcription factors associated with sox19a+sox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox19a
|
ENSDARG00000010770 | SRY-box transcription factor 19a |
|
sox2
|
ENSDARG00000070913 | SRY-box transcription factor 2 |
|
sox19a
|
ENSDARG00000110497 | SRY-box transcription factor 19a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox2 | dr11_v1_chr22_-_37349967_37349967 | 0.85 | 3.1e-27 | Click! |
| sox19a | dr11_v1_chr5_-_24201437_24201437 | 0.67 | 2.3e-13 | Click! |
Activity profile of sox19a+sox2 motif
Sorted Z-values of sox19a+sox2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_17745345 | 70.77 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr11_+_25735478 | 48.64 |
ENSDART00000103566
|
si:dkey-183j2.10
|
si:dkey-183j2.10 |
| chr6_-_43092175 | 43.92 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr11_+_35364445 | 41.88 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr14_-_33872092 | 41.67 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr2_-_21082695 | 41.52 |
ENSDART00000032502
|
nebl
|
nebulette |
| chr6_+_27146671 | 38.43 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr19_-_21832441 | 38.29 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr2_+_3472832 | 36.85 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr17_+_15535501 | 32.87 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr9_+_35901554 | 31.43 |
ENSDART00000005086
|
atp1a1b
|
ATPase Na+/K+ transporting subunit alpha 1b |
| chr7_-_24472991 | 31.36 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr4_-_27301356 | 31.19 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr7_-_26408472 | 30.62 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr8_+_16004551 | 30.15 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr19_+_43017628 | 29.45 |
ENSDART00000182608
ENSDART00000015632 |
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr16_+_34523515 | 29.40 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr24_-_33756003 | 29.35 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr16_+_25316973 | 29.08 |
ENSDART00000086409
|
dync1i1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr6_-_35487413 | 28.75 |
ENSDART00000102461
|
rgs8
|
regulator of G protein signaling 8 |
| chr9_+_42066030 | 28.36 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr16_+_14029283 | 27.64 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr21_+_13861589 | 27.08 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr16_-_27640995 | 26.39 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr20_+_35382482 | 26.27 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr5_-_6377865 | 25.63 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr24_+_25069609 | 25.60 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr9_+_2393764 | 25.29 |
ENSDART00000172624
|
chn1
|
chimerin 1 |
| chr16_-_6821927 | 25.19 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr25_-_204019 | 25.18 |
ENSDART00000188440
ENSDART00000191735 |
FP236318.2
|
|
| chr22_+_5106751 | 25.06 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr6_-_11780070 | 24.71 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr9_+_30108641 | 24.68 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr19_+_43017931 | 24.54 |
ENSDART00000132213
|
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr1_-_14233815 | 24.45 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr23_-_30787932 | 24.36 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr14_-_36378494 | 24.20 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr10_+_21783213 | 24.14 |
ENSDART00000168899
|
pcdh1g33
|
protocadherin 1 gamma 33 |
| chr9_-_18743012 | 24.12 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr23_-_21453614 | 23.97 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr19_-_19339285 | 23.78 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr13_+_51579851 | 23.71 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr20_+_18580176 | 23.36 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr21_+_26389391 | 23.22 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr10_+_21677058 | 23.20 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr9_-_18742704 | 22.94 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr6_+_3828560 | 22.52 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr12_+_33151246 | 22.32 |
ENSDART00000162681
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr2_-_31301929 | 22.29 |
ENSDART00000191992
ENSDART00000190723 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr3_-_22212764 | 22.23 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr2_+_26179096 | 22.16 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr23_-_18286822 | 22.09 |
ENSDART00000136672
|
fam19a1a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1a |
| chr9_-_44295071 | 21.94 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr24_-_27400017 | 21.87 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr14_-_1955257 | 21.49 |
ENSDART00000193254
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr8_+_16004154 | 21.48 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr14_-_32403554 | 21.02 |
ENSDART00000172873
ENSDART00000173408 ENSDART00000173114 ENSDART00000185594 ENSDART00000186762 ENSDART00000010982 |
fgf13a
|
fibroblast growth factor 13a |
| chr13_-_31441042 | 20.99 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr23_+_35708730 | 20.79 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr15_+_22311803 | 20.77 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr14_-_2361692 | 20.74 |
ENSDART00000167696
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.4 |
| chr14_-_2285955 | 20.60 |
ENSDART00000183928
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr14_-_1958994 | 20.58 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr7_+_14632157 | 20.47 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr2_+_47623202 | 20.36 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr15_+_32821392 | 20.34 |
ENSDART00000158272
|
dclk1b
|
doublecortin-like kinase 1b |
| chr21_-_42055872 | 20.32 |
ENSDART00000144767
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr8_+_39619087 | 20.31 |
ENSDART00000134822
|
msi1
|
musashi RNA-binding protein 1 |
| chr10_-_31782616 | 20.30 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr9_-_54840124 | 20.26 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr20_+_9828769 | 20.12 |
ENSDART00000160480
ENSDART00000053844 ENSDART00000080691 |
stxbp6l
|
syntaxin binding protein 6 (amisyn), like |
| chr14_-_2318590 | 19.81 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr14_-_2358774 | 19.73 |
ENSDART00000164809
|
si:ch73-233f7.5
|
si:ch73-233f7.5 |
| chr25_+_31929325 | 19.39 |
ENSDART00000181095
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr2_+_24177006 | 19.38 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr14_-_41285392 | 19.36 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr22_-_22416337 | 19.27 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr14_-_33872616 | 19.26 |
ENSDART00000162840
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr11_-_37548711 | 19.18 |
ENSDART00000127184
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr9_+_29585943 | 19.03 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr13_+_30506781 | 19.01 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr18_-_21218851 | 18.95 |
ENSDART00000060160
|
calb2a
|
calbindin 2a |
| chr5_+_37840914 | 18.89 |
ENSDART00000097738
|
panx1b
|
pannexin 1b |
| chr16_+_10318893 | 18.87 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr20_-_20821783 | 18.69 |
ENSDART00000152577
ENSDART00000027603 ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr17_+_23298928 | 18.63 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr20_+_30445971 | 18.63 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr5_+_64739762 | 18.52 |
ENSDART00000161112
ENSDART00000135610 ENSDART00000002908 |
olfm1a
|
olfactomedin 1a |
| chr7_+_23875269 | 18.51 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr5_-_33460959 | 18.25 |
ENSDART00000085636
|
si:ch211-182d3.1
|
si:ch211-182d3.1 |
| chr10_-_35542071 | 18.18 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr1_+_54911458 | 17.98 |
ENSDART00000089603
|
golga7ba
|
golgin A7 family, member Ba |
| chr1_-_45647846 | 17.97 |
ENSDART00000186881
|
BX511120.1
|
|
| chr25_+_37126921 | 17.86 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr21_+_13353263 | 17.81 |
ENSDART00000114677
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr14_-_2355833 | 17.72 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr23_-_21463788 | 17.58 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr9_+_27411502 | 17.57 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr23_-_21471022 | 17.55 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr19_+_12762887 | 17.54 |
ENSDART00000139909
|
mc5ra
|
melanocortin 5a receptor |
| chr11_+_26604224 | 17.49 |
ENSDART00000030453
ENSDART00000168895 ENSDART00000159505 |
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr23_+_23182037 | 17.44 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr16_-_12173554 | 17.41 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr20_+_41756996 | 17.39 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr4_+_3482312 | 17.27 |
ENSDART00000109044
|
grm8a
|
glutamate receptor, metabotropic 8a |
| chr20_+_37661229 | 17.24 |
ENSDART00000138539
|
aig1
|
androgen-induced 1 (H. sapiens) |
| chr2_-_5942115 | 17.09 |
ENSDART00000154489
|
tmem125b
|
transmembrane protein 125b |
| chr8_-_6877390 | 17.03 |
ENSDART00000170883
ENSDART00000005321 |
neflb
|
neurofilament, light polypeptide b |
| chr2_+_26303627 | 16.87 |
ENSDART00000040278
|
efna2a
|
ephrin-A2a |
| chr5_-_58939460 | 16.84 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr24_-_6644734 | 16.71 |
ENSDART00000167391
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr5_-_35301800 | 16.68 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr21_-_43043523 | 16.61 |
ENSDART00000040169
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr20_+_51312883 | 16.48 |
ENSDART00000084186
|
tmem151ba
|
transmembrane protein 151Ba |
| chr14_-_2364761 | 16.37 |
ENSDART00000167322
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.3 |
| chr20_-_48485354 | 16.36 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr14_-_1990290 | 16.32 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr14_-_33454595 | 16.31 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr4_+_12615836 | 16.27 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr20_-_27864964 | 16.25 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr10_+_39164638 | 15.89 |
ENSDART00000188997
|
CU633908.1
|
|
| chr14_-_2270973 | 15.89 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr13_-_43599898 | 15.87 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr15_-_16070731 | 15.84 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr13_+_25428677 | 15.69 |
ENSDART00000186284
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr7_-_38612230 | 15.68 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr17_-_15657029 | 15.53 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr16_+_5678071 | 15.41 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr17_-_44968177 | 15.39 |
ENSDART00000075510
|
ngb
|
neuroglobin |
| chr20_-_40319890 | 15.36 |
ENSDART00000075112
|
clvs2
|
clavesin 2 |
| chr23_-_14918276 | 15.28 |
ENSDART00000179831
|
ndrg3b
|
ndrg family member 3b |
| chr13_+_27951688 | 15.24 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr11_+_23704410 | 15.09 |
ENSDART00000112655
|
nfasca
|
neurofascin homolog (chicken) a |
| chr16_-_16590780 | 15.07 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr20_-_31497300 | 15.04 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr11_+_25101220 | 15.03 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr17_+_2063693 | 15.02 |
ENSDART00000182349
|
zgc:162989
|
zgc:162989 |
| chr9_-_18735256 | 14.91 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr3_+_24134418 | 14.70 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr15_-_33925851 | 14.70 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr7_-_52842007 | 14.65 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr2_+_26240631 | 14.60 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr10_-_22845485 | 14.60 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr7_+_49078890 | 14.59 |
ENSDART00000067857
|
b4galnt4b
|
beta-1,4-N-acetyl-galactosaminyl transferase 4b |
| chr5_-_26093945 | 14.52 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr10_+_18952271 | 14.51 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr13_-_27660955 | 14.31 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr17_+_6793001 | 14.30 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr19_+_30633453 | 14.29 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr21_+_17110598 | 14.27 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr15_+_36115955 | 14.23 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr19_+_16032383 | 14.19 |
ENSDART00000046530
|
rab42a
|
RAB42, member RAS oncogene family a |
| chr16_-_15988320 | 14.14 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr4_-_14315855 | 14.12 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr2_+_24177190 | 13.99 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr8_+_25267903 | 13.98 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr18_-_38088099 | 13.89 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr19_-_103289 | 13.88 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr5_-_32323136 | 13.87 |
ENSDART00000110804
|
hspb15
|
heat shock protein, alpha-crystallin-related, b15 |
| chr10_+_21559605 | 13.87 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr4_-_24031924 | 13.85 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr14_-_26177156 | 13.83 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr11_+_40649412 | 13.82 |
ENSDART00000043016
ENSDART00000134560 |
slc45a1
|
solute carrier family 45, member 1 |
| chr2_-_32688905 | 13.78 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr17_+_8183393 | 13.75 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr2_-_21170517 | 13.74 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr5_+_23118470 | 13.74 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr18_+_642889 | 13.73 |
ENSDART00000189007
|
CABZ01078320.1
|
|
| chr17_-_16965809 | 13.72 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr12_+_10115964 | 13.63 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr13_+_12739283 | 13.61 |
ENSDART00000102279
|
lingo2b
|
leucine rich repeat and Ig domain containing 2b |
| chr1_-_25911901 | 13.61 |
ENSDART00000109714
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr21_+_45733871 | 13.59 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr15_-_33896159 | 13.59 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr17_+_12700617 | 13.53 |
ENSDART00000191931
|
stmn4l
|
stathmin-like 4, like |
| chr22_-_20309283 | 13.52 |
ENSDART00000182125
ENSDART00000048775 |
si:dkey-110c1.10
|
si:dkey-110c1.10 |
| chr10_+_21668606 | 13.48 |
ENSDART00000185751
|
CT737184.2
|
|
| chr5_-_29488245 | 13.46 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr6_-_30210378 | 13.42 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr18_-_12957451 | 13.40 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr2_+_34767171 | 13.37 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr1_-_31505144 | 13.35 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr16_+_2840775 | 13.35 |
ENSDART00000108735
|
clec3ba
|
C-type lectin domain family 3, member Ba |
| chr25_+_21833287 | 13.35 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr11_+_24966182 | 13.33 |
ENSDART00000185088
|
sulf2a
|
sulfatase 2a |
| chr23_-_12015139 | 13.28 |
ENSDART00000110627
ENSDART00000193988 ENSDART00000184528 |
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr16_-_12953739 | 13.26 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr17_-_36936649 | 13.23 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr4_-_11163112 | 13.17 |
ENSDART00000188854
|
prmt8b
|
protein arginine methyltransferase 8b |
| chr14_-_2322484 | 13.13 |
ENSDART00000167806
|
si:ch73-379j16.2
|
si:ch73-379j16.2 |
| chr5_+_65491390 | 13.10 |
ENSDART00000159921
|
si:dkey-21e5.1
|
si:dkey-21e5.1 |
| chr2_+_19777146 | 13.09 |
ENSDART00000038648
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr16_+_33593116 | 13.05 |
ENSDART00000013148
|
pou3f1
|
POU class 3 homeobox 1 |
| chr4_-_64284924 | 12.97 |
ENSDART00000166733
|
CT956002.2
|
|
| chr25_-_7925269 | 12.94 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr10_-_22249444 | 12.93 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr20_-_26042070 | 12.92 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr2_+_20430366 | 12.91 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr8_-_52473054 | 12.87 |
ENSDART00000193658
|
phyhip
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr23_-_36449111 | 12.80 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox19a+sox2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 7.3 | 21.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 7.2 | 28.7 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 6.8 | 20.4 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 6.8 | 20.3 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 6.3 | 25.3 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 5.7 | 23.0 | GO:0016123 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 5.5 | 43.9 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 5.2 | 20.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 5.0 | 15.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 4.7 | 18.9 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 4.1 | 20.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 4.1 | 36.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 4.0 | 12.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 3.8 | 11.5 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 3.7 | 32.9 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 3.5 | 13.9 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 3.4 | 13.7 | GO:2000048 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 3.4 | 10.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 3.4 | 10.2 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 3.2 | 73.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 3.2 | 47.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 3.1 | 12.6 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 2.9 | 8.8 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 2.9 | 8.7 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 2.9 | 11.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 2.9 | 22.9 | GO:0021634 | optic nerve formation(GO:0021634) |
| 2.8 | 39.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 2.8 | 11.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 2.8 | 19.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 2.8 | 33.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 2.7 | 18.9 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 2.7 | 8.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 2.6 | 36.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 2.6 | 23.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 2.5 | 15.3 | GO:0022029 | telencephalon cell migration(GO:0022029) |
| 2.5 | 22.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 2.5 | 32.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 2.5 | 39.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 2.4 | 7.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 2.4 | 23.8 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 2.4 | 7.1 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 2.3 | 25.5 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 2.3 | 9.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 2.3 | 16.1 | GO:0019857 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 2.2 | 6.7 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 2.2 | 26.6 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 2.2 | 26.4 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 2.2 | 8.7 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 2.2 | 19.4 | GO:0001840 | neural plate development(GO:0001840) |
| 2.1 | 10.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 2.1 | 14.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 2.1 | 12.6 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 2.1 | 8.4 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 2.1 | 18.5 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 2.0 | 22.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 1.9 | 23.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 1.9 | 7.6 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 1.9 | 7.5 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 1.9 | 11.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 1.9 | 7.5 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 1.8 | 5.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.8 | 9.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.8 | 8.9 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 1.8 | 447.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 1.7 | 17.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 1.7 | 13.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 1.7 | 22.2 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 1.7 | 8.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 1.7 | 5.0 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 1.7 | 5.0 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 1.7 | 31.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 1.7 | 8.3 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 1.6 | 14.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 1.6 | 6.6 | GO:0035521 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.6 | 57.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 1.6 | 61.1 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 1.6 | 16.0 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 1.6 | 8.0 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 1.6 | 68.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 1.6 | 22.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 1.6 | 3.1 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 1.6 | 34.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 1.5 | 4.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 1.5 | 9.2 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 1.5 | 27.7 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 1.5 | 4.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.5 | 17.9 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 1.5 | 4.4 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 1.5 | 27.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 1.5 | 10.2 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 1.4 | 20.1 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 1.4 | 4.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.4 | 9.8 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 1.4 | 5.6 | GO:0010226 | response to lithium ion(GO:0010226) |
| 1.4 | 8.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 1.3 | 8.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.3 | 5.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.3 | 17.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 1.3 | 11.7 | GO:0001780 | neutrophil homeostasis(GO:0001780) response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.3 | 6.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 1.3 | 42.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 1.3 | 46.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 1.2 | 21.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 1.2 | 2.5 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 1.2 | 9.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 1.2 | 10.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.2 | 11.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 1.2 | 11.6 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 1.2 | 4.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 1.1 | 79.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 1.1 | 18.3 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 1.1 | 12.5 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 1.1 | 12.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 1.1 | 14.6 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 1.1 | 15.6 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 1.1 | 5.5 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 1.1 | 15.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 1.1 | 7.7 | GO:0021982 | pineal gland development(GO:0021982) |
| 1.1 | 5.5 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 1.1 | 36.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 1.1 | 5.4 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 1.1 | 10.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 1.1 | 10.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 1.0 | 3.1 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 1.0 | 2.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 1.0 | 3.1 | GO:0060959 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 1.0 | 16.2 | GO:0036065 | fucosylation(GO:0036065) |
| 1.0 | 26.1 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 1.0 | 14.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.0 | 8.9 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 1.0 | 4.9 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.9 | 6.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.9 | 29.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.9 | 8.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.9 | 6.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.9 | 3.6 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.9 | 4.5 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.9 | 1.8 | GO:0007021 | tubulin complex assembly(GO:0007021) post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.9 | 4.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.9 | 2.7 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.9 | 4.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.9 | 13.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.9 | 4.3 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.9 | 5.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.8 | 4.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.8 | 2.5 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.8 | 5.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.8 | 21.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.8 | 10.8 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.8 | 20.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.8 | 11.5 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.8 | 5.7 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.8 | 4.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.8 | 7.3 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.8 | 8.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.8 | 32.2 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.8 | 15.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.8 | 7.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.8 | 22.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.8 | 3.9 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.8 | 27.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.8 | 1.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.8 | 8.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.8 | 4.5 | GO:1903826 | arginine transport(GO:0015809) L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.8 | 2.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.8 | 9.8 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.7 | 7.4 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.7 | 7.2 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.7 | 5.7 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.7 | 12.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.7 | 1.4 | GO:0006984 | ER-nucleus signaling pathway(GO:0006984) |
| 0.7 | 42.7 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.7 | 5.6 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.7 | 9.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.7 | 2.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.7 | 4.8 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.7 | 5.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.7 | 5.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.7 | 4.7 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.7 | 12.0 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.7 | 35.5 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.6 | 5.8 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.6 | 5.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.6 | 2.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.6 | 19.9 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.6 | 8.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.6 | 27.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.6 | 9.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.6 | 3.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.6 | 7.4 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.6 | 13.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.6 | 6.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.6 | 1.2 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.6 | 3.6 | GO:0035313 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.6 | 7.7 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.6 | 5.3 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.6 | 2.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.6 | 5.7 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.6 | 2.9 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.6 | 8.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.6 | 1.7 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.6 | 6.7 | GO:0060384 | innervation(GO:0060384) axonogenesis involved in innervation(GO:0060385) |
| 0.6 | 11.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.6 | 11.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.6 | 5.0 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.6 | 2.8 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.5 | 3.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.5 | 8.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.5 | 3.8 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.5 | 1.6 | GO:0071922 | regulation of sister chromatid cohesion(GO:0007063) regulation of chromosome condensation(GO:0060623) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.5 | 3.8 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.5 | 11.8 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.5 | 2.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.5 | 5.8 | GO:0072176 | pronephric duct development(GO:0039022) nephric duct development(GO:0072176) |
| 0.5 | 2.6 | GO:0051103 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.5 | 6.3 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.5 | 14.5 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.5 | 8.8 | GO:1900151 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) positive regulation of mRNA catabolic process(GO:0061014) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.5 | 1.5 | GO:0042762 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of sulfur metabolic process(GO:0042762) regulation of homocysteine metabolic process(GO:0050666) |
| 0.5 | 14.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.5 | 6.1 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.5 | 6.1 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.5 | 1.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.5 | 2.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.5 | 9.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.5 | 44.6 | GO:0010970 | establishment of localization by movement along microtubule(GO:0010970) |
| 0.5 | 11.4 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.5 | 10.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.5 | 37.1 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.5 | 5.3 | GO:0072160 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.5 | 48.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.5 | 4.8 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.5 | 3.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.5 | 2.4 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.5 | 9.5 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 3.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.5 | 2.3 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.5 | 89.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.5 | 4.6 | GO:0035907 | dorsal aorta development(GO:0035907) |
| 0.5 | 1.4 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.5 | 2.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.5 | 1.4 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.5 | 4.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.4 | 2.7 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.4 | 13.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.4 | 54.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.4 | 15.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.4 | 1.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.4 | 2.6 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.4 | 2.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.4 | 2.5 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.4 | 1.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 10.9 | GO:0071696 | ectodermal placode development(GO:0071696) |
| 0.4 | 17.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.4 | 5.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.4 | 4.6 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.4 | 6.9 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.4 | 1.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.4 | 1.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 3.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 27.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.4 | 4.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.4 | 1.5 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.4 | 2.3 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.4 | 43.2 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.4 | 8.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.4 | 2.2 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.4 | 7.7 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.4 | 1.1 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.4 | 14.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.4 | 15.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.4 | 1.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.3 | 3.8 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 3.7 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.3 | 1.4 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.3 | 11.9 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.3 | 3.0 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.3 | 1.0 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.3 | 18.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.3 | 26.4 | GO:0006475 | internal protein amino acid acetylation(GO:0006475) internal peptidyl-lysine acetylation(GO:0018393) |
| 0.3 | 2.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 6.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 3.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 6.8 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 57.1 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.3 | 1.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 17.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 7.4 | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation(GO:0010770) |
| 0.3 | 7.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.3 | 1.9 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 11.9 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.3 | 2.4 | GO:0060021 | palate development(GO:0060021) |
| 0.3 | 1.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 11.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.3 | 6.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.3 | 8.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.3 | 1.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) peptidyl-lysine dimethylation(GO:0018027) |
| 0.3 | 64.1 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.3 | 12.4 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.3 | 4.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.3 | 27.5 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.3 | 2.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 11.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.3 | 6.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 2.2 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.3 | 4.4 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.3 | 4.9 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.3 | 4.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.3 | 0.5 | GO:0046324 | regulation of glucose transport(GO:0010827) regulation of glucose import(GO:0046324) |
| 0.3 | 1.0 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.3 | 6.2 | GO:1902275 | regulation of chromatin organization(GO:1902275) |
| 0.3 | 5.6 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.3 | 3.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 3.2 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.2 | 0.5 | GO:2000142 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.2 | 4.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 3.1 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.2 | 5.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 3.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 2.8 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 5.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 3.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 0.2 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 0.7 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 124.6 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.2 | 3.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 1.4 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 1.5 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 7.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.2 | 5.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.2 | 13.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 4.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 9.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.2 | 2.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 3.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.2 | 2.9 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 7.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.2 | 5.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.2 | 2.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 3.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 22.4 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.2 | 0.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 4.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.2 | 3.0 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.2 | 5.3 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.2 | 68.9 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.2 | 3.9 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.2 | 2.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 9.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 1.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.2 | 1.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 1.6 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 12.2 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.2 | 11.2 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.2 | 1.4 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.2 | 2.1 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 1.3 | GO:0032196 | transposition(GO:0032196) |
| 0.2 | 2.0 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.2 | 10.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.2 | 4.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 15.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.2 | 1.1 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.6 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 2.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 16.3 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.1 | 0.4 | GO:1901255 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 7.2 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 4.0 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 5.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 1.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 1.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 7.5 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.1 | 0.6 | GO:0014004 | microglia differentiation(GO:0014004) |
| 0.1 | 2.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 11.3 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.1 | 1.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) |
| 0.1 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.5 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 4.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 3.1 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.1 | 24.8 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.1 | 1.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 6.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 1.4 | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.1 | 0.8 | GO:1903321 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.1 | 3.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.9 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.1 | 1.1 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 3.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 2.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.4 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.1 | 1.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 1.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.2 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.1 | 0.6 | GO:0046512 | sphingosine metabolic process(GO:0006670) diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.9 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 1.2 | GO:0045664 | regulation of neuron differentiation(GO:0045664) |
| 0.0 | 3.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0034724 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.2 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.4 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 9.4 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
| 0.0 | 3.6 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0060996 | dendritic spine development(GO:0060996) dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 3.9 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.0 | 0.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 53.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 4.4 | 26.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 4.4 | 13.2 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 4.1 | 73.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 3.6 | 151.2 | GO:0043204 | perikaryon(GO:0043204) |
| 3.5 | 48.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 3.4 | 17.0 | GO:0005883 | neurofilament(GO:0005883) |
| 3.4 | 3.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 3.3 | 20.0 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 3.2 | 22.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 3.0 | 33.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 2.8 | 8.5 | GO:0031213 | RSF complex(GO:0031213) |
| 2.6 | 7.8 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 2.5 | 12.5 | GO:0008091 | spectrin(GO:0008091) |
| 2.3 | 23.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 2.3 | 25.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.8 | 42.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 1.8 | 70.8 | GO:0043679 | axon terminus(GO:0043679) |
| 1.7 | 17.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 1.7 | 5.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.7 | 40.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 1.6 | 4.7 | GO:1990745 | EARP complex(GO:1990745) |
| 1.5 | 4.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 1.5 | 17.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 1.5 | 27.7 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 1.4 | 5.6 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 1.4 | 46.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.2 | 8.7 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 1.2 | 22.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 1.2 | 18.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 1.2 | 32.5 | GO:0000145 | exocyst(GO:0000145) |
| 1.1 | 8.0 | GO:0033010 | paranodal junction(GO:0033010) |
| 1.1 | 4.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.1 | 16.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.1 | 4.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 1.1 | 9.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.1 | 4.2 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 1.0 | 11.3 | GO:0035101 | FACT complex(GO:0035101) |
| 1.0 | 18.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 1.0 | 23.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 1.0 | 4.0 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 1.0 | 3.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.0 | 15.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.9 | 32.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.9 | 3.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.9 | 2.7 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.9 | 5.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.8 | 4.2 | GO:0000938 | GARP complex(GO:0000938) |
| 0.8 | 43.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.8 | 3.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.8 | 19.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.8 | 2.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.8 | 5.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.7 | 41.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.7 | 2.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.7 | 5.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.7 | 27.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.7 | 8.7 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.7 | 18.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.7 | 7.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.7 | 68.0 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.7 | 6.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.7 | 6.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.7 | 48.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.6 | 13.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.6 | 8.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.6 | 110.3 | GO:0030424 | axon(GO:0030424) |
| 0.6 | 7.5 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.6 | 16.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.6 | 24.9 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.6 | 2.9 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.6 | 8.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.6 | 2.9 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.6 | 5.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.6 | 10.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.6 | 2.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 11.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.5 | 3.8 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.5 | 32.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.5 | 51.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.5 | 15.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.5 | 7.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.5 | 13.2 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.5 | 7.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.5 | 206.2 | GO:0043005 | neuron projection(GO:0043005) |
| 0.5 | 8.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.5 | 3.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.5 | 3.9 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.5 | 6.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.5 | 5.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.5 | 8.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.5 | 3.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.4 | 2.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.4 | 38.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.4 | 19.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.4 | 1.7 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.4 | 1.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.4 | 15.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.4 | 8.2 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.4 | 4.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 2.8 | GO:0016586 | RSC complex(GO:0016586) |
| 0.4 | 12.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.4 | 24.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 5.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 8.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.4 | 3.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.4 | 2.7 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.4 | 28.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.4 | 2.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.4 | 6.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.4 | 1.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.4 | 2.6 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.4 | 1.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 22.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.3 | 8.9 | GO:0030496 | midbody(GO:0030496) |
| 0.3 | 10.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.3 | 2.7 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.3 | 5.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 2.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 1.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 3.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.3 | 4.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.3 | 8.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.3 | 9.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.3 | 6.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.3 | 25.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.3 | 335.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.3 | 9.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 1.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 2.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 82.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.2 | 13.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.2 | 13.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 5.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 1.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 2.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 26.7 | GO:0045202 | synapse(GO:0045202) |
| 0.2 | 8.3 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.2 | 14.2 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.2 | 52.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.2 | 3.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 2.0 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 13.2 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.2 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 1.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 8.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 42.0 | GO:0097708 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
| 0.2 | 2.3 | GO:0098858 | actin-based cell projection(GO:0098858) |
| 0.2 | 8.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 2.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.2 | 2.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 7.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.8 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 2.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 18.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 8.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 3.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 13.7 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.1 | 8.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 0.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 27.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.1 | 2.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.5 | GO:0070938 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.1 | 6.9 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 397.3 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 6.3 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 2.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 85.8 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 1.9 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 53.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 7.5 | 22.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 6.4 | 25.7 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 5.9 | 23.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 5.7 | 23.0 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 5.6 | 22.3 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 3.9 | 11.7 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 3.9 | 58.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 3.8 | 15.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 3.8 | 15.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 3.4 | 20.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 3.3 | 39.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 3.1 | 31.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 3.1 | 15.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 3.1 | 9.2 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 3.1 | 12.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 3.0 | 21.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 3.0 | 42.0 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 3.0 | 27.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 3.0 | 26.6 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 2.8 | 19.6 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 2.7 | 13.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 2.5 | 14.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 2.5 | 17.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 2.4 | 31.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 2.4 | 7.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 2.3 | 25.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 2.3 | 16.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 2.2 | 29.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 2.2 | 11.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 2.2 | 17.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 2.1 | 19.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 2.1 | 12.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.0 | 4.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 2.0 | 22.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 2.0 | 18.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.9 | 13.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 1.9 | 89.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 1.8 | 5.5 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 1.7 | 10.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 1.7 | 22.2 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 1.7 | 22.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 1.7 | 11.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.7 | 15.0 | GO:0016934 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.6 | 31.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 1.6 | 37.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 1.6 | 28.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.5 | 9.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 1.5 | 7.5 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 1.4 | 54.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.3 | 8.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 1.3 | 28.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 1.3 | 10.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 1.3 | 5.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 1.3 | 14.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.3 | 30.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 1.2 | 7.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 1.2 | 33.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.2 | 17.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 1.2 | 5.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 1.2 | 20.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.1 | 37.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.1 | 4.6 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 1.1 | 4.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.1 | 9.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.1 | 6.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 1.1 | 5.5 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 1.1 | 6.4 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 1.1 | 9.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 1.0 | 13.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 1.0 | 3.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 1.0 | 20.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 1.0 | 3.1 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 1.0 | 7.2 | GO:0001032 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 1.0 | 3.0 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 1.0 | 5.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 1.0 | 8.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.0 | 6.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 1.0 | 7.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 1.0 | 44.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 1.0 | 17.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 1.0 | 16.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.9 | 4.7 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.9 | 8.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.9 | 3.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.9 | 3.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.9 | 4.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.9 | 10.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.9 | 9.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.9 | 4.5 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.9 | 5.4 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.9 | 5.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.9 | 4.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.9 | 14.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.9 | 19.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.9 | 2.6 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.9 | 42.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.9 | 11.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.9 | 2.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.9 | 6.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.8 | 6.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.8 | 21.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.8 | 4.7 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.8 | 25.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.8 | 12.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.8 | 10.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.8 | 6.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.7 | 4.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.7 | 12.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 3.5 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.7 | 5.6 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.7 | 4.9 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.7 | 3.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.7 | 5.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.7 | 9.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.7 | 24.6 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.7 | 11.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.7 | 5.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.7 | 2.0 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.7 | 10.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.7 | 2.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.7 | 6.6 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 3.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 15.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.6 | 4.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.6 | 14.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.6 | 243.8 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.6 | 9.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.6 | 7.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.6 | 3.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.6 | 8.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 8.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.6 | 7.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.6 | 4.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.6 | 4.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.6 | 8.9 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.6 | 8.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.6 | 27.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.6 | 10.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.6 | 2.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.6 | 3.5 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.6 | 22.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.6 | 6.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.6 | 11.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.6 | 7.4 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.6 | 5.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.6 | 3.9 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.6 | 11.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 5.0 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.5 | 8.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.5 | 2.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.5 | 45.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.5 | 60.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.5 | 3.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.5 | 3.5 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.5 | 2.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.5 | 19.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.5 | 3.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.5 | 3.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.5 | 18.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.5 | 14.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 20.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.5 | 6.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.5 | 14.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 1.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.5 | 1.4 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.4 | 7.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.4 | 8.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.4 | 1.8 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 3.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.4 | 6.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.4 | 420.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.4 | 16.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.4 | 3.0 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.4 | 1.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.4 | 3.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 16.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.4 | 4.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.4 | 2.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.4 | 6.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.4 | 17.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.4 | 134.1 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.4 | 8.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.4 | 9.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.4 | 9.4 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.4 | 2.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.4 | 1.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.4 | 2.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.3 | 4.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 1.7 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.3 | 8.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 17.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.3 | 2.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 5.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.3 | 6.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.3 | 1.6 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.3 | 2.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 1.3 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.3 | 6.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 1.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.3 | 27.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.3 | 4.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 1.5 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.3 | 7.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.3 | 7.0 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.3 | 6.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 1.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.3 | 11.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 0.8 | GO:0032183 | SUMO binding(GO:0032183) SUMO polymer binding(GO:0032184) |
| 0.3 | 1.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 1.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 1.1 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.3 | 23.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.3 | 107.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 6.6 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.3 | 42.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.3 | 93.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.3 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 30.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.3 | 4.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.3 | 3.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 5.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 7.6 | GO:0045502 | dynein binding(GO:0045502) |
| 0.2 | 8.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 11.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.2 | 2.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 2.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 2.1 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 5.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 3.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.2 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 48.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.2 | 4.1 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 9.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 0.6 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.2 | 1.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 0.6 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 4.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 4.5 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.2 | 7.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 29.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 0.9 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 25.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.2 | 5.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.2 | 3.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.2 | 3.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 5.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 1.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 4.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 7.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 9.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 2.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 253.0 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 5.5 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 16.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 5.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 6.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 4.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.5 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 1.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.4 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 1.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 4.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 7.4 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 0.5 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 15.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 1.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 13.9 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 1.9 | 40.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 1.3 | 11.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 1.2 | 8.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 1.1 | 36.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 1.1 | 42.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 1.1 | 17.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.9 | 27.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.9 | 7.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.7 | 2.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.6 | 16.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.5 | 4.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 6.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.5 | 24.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.4 | 7.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.4 | 2.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.4 | 1.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 7.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.4 | 15.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.4 | 3.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.4 | 11.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.3 | 5.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.3 | 7.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 1.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.3 | 6.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.3 | 9.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.3 | 4.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.3 | 2.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 2.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 3.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 7.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 2.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 3.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 3.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 6.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 4.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 3.7 | 11.1 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 2.3 | 9.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 2.1 | 32.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.6 | 6.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 1.5 | 7.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 1.4 | 31.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 1.3 | 20.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 1.2 | 26.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 1.2 | 21.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 1.1 | 11.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 1.1 | 8.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 1.1 | 8.8 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.9 | 24.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.9 | 15.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.8 | 13.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.8 | 13.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 12.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.8 | 8.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.8 | 3.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.8 | 4.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.7 | 5.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.7 | 5.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.7 | 6.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.7 | 6.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 6.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.6 | 14.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.6 | 7.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.6 | 7.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.5 | 2.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 9.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.5 | 4.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 8.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.4 | 3.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.4 | 4.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.4 | 5.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.4 | 5.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 22.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.3 | 3.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.3 | 9.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.3 | 10.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.3 | 4.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 2.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.3 | 3.4 | REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.3 | 1.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.3 | 4.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 3.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 1.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 5.4 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.2 | 2.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 2.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 7.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 2.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 2.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 4.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 11.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.1 | 2.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 14.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.2 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 3.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 2.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 3.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |