Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for si:dkey-18a10.3
Z-value: 1.21
Transcription factors associated with si:dkey-18a10.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-18a10.3
|
ENSDARG00000090814 | si_dkey-18a10.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-18a10.3 | dr11_v1_chr10_-_33572441_33572441 | -0.28 | 6.3e-03 | Click! |
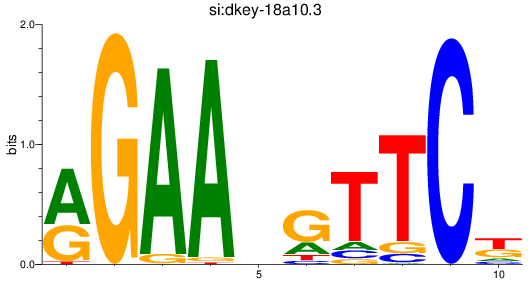
Activity profile of si:dkey-18a10.3 motif
Sorted Z-values of si:dkey-18a10.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_53996193 | 22.65 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr21_+_20383837 | 14.91 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr13_+_24834199 | 14.55 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr15_+_6109861 | 12.05 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr9_+_44431174 | 12.04 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr16_+_34523515 | 11.80 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr16_-_6821927 | 9.41 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr7_+_29955368 | 9.38 |
ENSDART00000173686
|
tpma
|
alpha-tropomyosin |
| chr7_+_29954709 | 9.30 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr15_-_37850969 | 9.20 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr23_-_46217134 | 8.41 |
ENSDART00000189477
ENSDART00000168352 |
CABZ01102528.1
|
|
| chr5_-_20879527 | 7.78 |
ENSDART00000134697
|
pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr23_-_31372639 | 7.60 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr9_+_44430974 | 7.34 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr19_-_35596207 | 6.88 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr22_+_18886209 | 6.79 |
ENSDART00000144402
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr4_-_16353733 | 6.54 |
ENSDART00000186785
|
lum
|
lumican |
| chr4_+_8168514 | 6.39 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr20_-_44496245 | 6.09 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr1_+_31054915 | 5.55 |
ENSDART00000148968
|
itga6b
|
integrin, alpha 6b |
| chr22_+_5574952 | 5.49 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr13_-_23007813 | 5.39 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr5_-_13685047 | 5.31 |
ENSDART00000018351
|
zgc:65851
|
zgc:65851 |
| chr4_-_789645 | 5.17 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr10_+_17936441 | 5.16 |
ENSDART00000146489
ENSDART00000064866 |
prkab1a
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, a |
| chr15_+_46357080 | 5.00 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr13_-_45475289 | 4.82 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr8_-_24970790 | 4.75 |
ENSDART00000141267
|
si:ch211-199o1.2
|
si:ch211-199o1.2 |
| chr19_+_46259619 | 4.66 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr5_+_32791245 | 4.62 |
ENSDART00000077189
|
ier5l
|
immediate early response 5-like |
| chr8_+_22931427 | 4.31 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr6_-_40697585 | 4.30 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr9_+_44430705 | 4.27 |
ENSDART00000190696
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr6_-_60147517 | 4.24 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr9_-_18735256 | 4.12 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr15_+_36941490 | 4.07 |
ENSDART00000172664
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr6_+_46341306 | 4.07 |
ENSDART00000111905
|
BX649498.1
|
|
| chr4_+_10616626 | 4.01 |
ENSDART00000067251
ENSDART00000143690 |
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr7_+_1467863 | 3.94 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr10_+_18952271 | 3.90 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr12_-_35787801 | 3.85 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr3_-_59981476 | 3.82 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr19_-_9712530 | 3.77 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr13_+_30506781 | 3.63 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr1_-_55750208 | 3.59 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr25_+_19999623 | 3.58 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr12_+_45200744 | 3.55 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr13_-_31452516 | 3.55 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr25_-_14433503 | 3.48 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr20_-_49657134 | 3.40 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr17_-_31044803 | 3.38 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr11_-_2456656 | 3.34 |
ENSDART00000171219
ENSDART00000167938 |
slc2a10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr9_-_56272465 | 3.21 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_+_33345348 | 3.20 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr1_-_1631399 | 3.20 |
ENSDART00000176787
|
clic6
|
chloride intracellular channel 6 |
| chr23_-_35066816 | 3.08 |
ENSDART00000168731
ENSDART00000163731 |
BX294434.1
|
|
| chr13_-_13751017 | 3.03 |
ENSDART00000180182
|
ky
|
kyphoscoliosis peptidase |
| chr2_-_35566938 | 2.92 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr16_-_13662514 | 2.91 |
ENSDART00000146348
|
shisa7a
|
shisa family member 7a |
| chr14_-_17622080 | 2.88 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr4_+_33012407 | 2.86 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr1_+_2431956 | 2.86 |
ENSDART00000183832
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr20_+_37393134 | 2.84 |
ENSDART00000128321
|
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr6_-_57655299 | 2.83 |
ENSDART00000083807
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr19_+_19767567 | 2.72 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr2_+_36015049 | 2.72 |
ENSDART00000158276
|
lamc2
|
laminin, gamma 2 |
| chr25_+_35942867 | 2.71 |
ENSDART00000066985
|
hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr6_+_40723554 | 2.70 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr20_+_23498255 | 2.69 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr17_-_42213285 | 2.68 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr17_+_41463942 | 2.62 |
ENSDART00000075331
|
insm1b
|
insulinoma-associated 1b |
| chr14_+_36223097 | 2.58 |
ENSDART00000186872
|
pitx2
|
paired-like homeodomain 2 |
| chr19_+_20178978 | 2.54 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr10_+_21776911 | 2.53 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr9_-_1951144 | 2.52 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr19_+_3842891 | 2.48 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr5_-_30176970 | 2.48 |
ENSDART00000098300
|
adamts8a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8a |
| chr25_+_35250976 | 2.43 |
ENSDART00000003494
|
slc17a6a
|
solute carrier family 17 (vesicular glutamate transporter), member 6a |
| chr2_+_24199276 | 2.43 |
ENSDART00000140575
|
map4l
|
microtubule associated protein 4 like |
| chr11_+_10548171 | 2.40 |
ENSDART00000191497
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr24_+_21973929 | 2.39 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr8_-_1909840 | 2.39 |
ENSDART00000147408
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr2_+_31957554 | 2.37 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr5_-_13778726 | 2.36 |
ENSDART00000051655
|
snrnp27
|
small nuclear ribonucleoprotein 27 (U4/U6.U5) |
| chr7_-_20103384 | 2.35 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr5_+_41496490 | 2.35 |
ENSDART00000039369
|
fancg
|
Fanconi anemia, complementation group G |
| chr15_+_28119284 | 2.34 |
ENSDART00000188366
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr20_+_36820965 | 2.28 |
ENSDART00000153085
ENSDART00000062935 |
heca
|
hdc homolog, cell cycle regulator |
| chr18_+_6536598 | 2.27 |
ENSDART00000149350
|
fkbp4
|
FK506 binding protein 4 |
| chr1_-_681116 | 2.24 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr20_+_23501535 | 2.23 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr25_-_8030113 | 2.19 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr5_-_30475011 | 2.16 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr17_+_21514777 | 2.14 |
ENSDART00000154633
|
chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr18_+_18455029 | 2.14 |
ENSDART00000165079
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr9_-_327901 | 2.12 |
ENSDART00000159956
|
ndufa4l2a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2a |
| chr5_+_31811662 | 2.11 |
ENSDART00000023463
|
uap1l1
|
UDP-N-acetylglucosamine pyrophosphorylase 1, like 1 |
| chr8_-_13364879 | 2.10 |
ENSDART00000063863
|
ccdc124
|
coiled-coil domain containing 124 |
| chr5_+_20103085 | 2.09 |
ENSDART00000079424
|
si:rp71-1c23.3
|
si:rp71-1c23.3 |
| chr6_+_49412754 | 2.07 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr9_+_32301017 | 2.06 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr9_+_32301456 | 2.05 |
ENSDART00000078608
ENSDART00000185153 ENSDART00000144947 |
hspe1
|
heat shock 10 protein 1 |
| chr10_+_17875891 | 2.05 |
ENSDART00000191744
|
phf24
|
PHD finger protein 24 |
| chr24_+_20969358 | 2.04 |
ENSDART00000129675
|
drd3
|
dopamine receptor D3 |
| chr2_-_35103069 | 2.00 |
ENSDART00000111730
|
pappa2
|
pappalysin 2 |
| chr6_-_24392909 | 1.99 |
ENSDART00000171042
ENSDART00000168355 |
brdt
|
bromodomain, testis-specific |
| chr6_-_13401133 | 1.98 |
ENSDART00000151206
|
fmnl2b
|
formin-like 2b |
| chr12_+_19138452 | 1.94 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr3_-_5067585 | 1.90 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr23_-_44494401 | 1.88 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr4_-_43822179 | 1.88 |
ENSDART00000185246
|
si:dkey-261p22.1
|
si:dkey-261p22.1 |
| chr2_-_41620112 | 1.84 |
ENSDART00000138822
ENSDART00000004816 |
zgc:110158
|
zgc:110158 |
| chr2_+_36616830 | 1.84 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr6_+_202367 | 1.81 |
ENSDART00000168232
|
mgat3b
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase b |
| chr19_+_35799384 | 1.80 |
ENSDART00000076023
|
angpt2b
|
angiopoietin 2b |
| chr17_-_44584811 | 1.78 |
ENSDART00000165059
ENSDART00000165252 |
slc35f4
|
solute carrier family 35, member F4 |
| chr13_-_46882267 | 1.77 |
ENSDART00000161322
|
slc29a1a
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1a |
| chr4_-_24019711 | 1.76 |
ENSDART00000077926
|
celf2
|
cugbp, Elav-like family member 2 |
| chr1_-_59411901 | 1.69 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr14_+_25817246 | 1.64 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr5_-_23999777 | 1.59 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr16_+_41517188 | 1.58 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr3_+_26267725 | 1.55 |
ENSDART00000131288
|
adap2
|
ArfGAP with dual PH domains 2 |
| chr15_-_22147860 | 1.55 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr8_+_14778292 | 1.54 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr1_+_45056371 | 1.53 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr6_-_24392426 | 1.47 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr19_+_12237945 | 1.43 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr3_-_58650057 | 1.43 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr23_-_24682244 | 1.43 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr2_+_30103131 | 1.41 |
ENSDART00000185119
|
dnajb6a
|
DnaJ (Hsp40) homolog, subfamily B, member 6a |
| chr10_+_1052591 | 1.38 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr21_-_37027252 | 1.38 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr18_+_14477740 | 1.36 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr16_+_21330634 | 1.36 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr16_-_32221312 | 1.32 |
ENSDART00000112348
|
calhm5.1
|
calcium homeostasis modulator family member 5, tandem duplicate 1 |
| chr6_-_35032792 | 1.31 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr3_-_36612877 | 1.30 |
ENSDART00000167164
|
si:dkeyp-72e1.7
|
si:dkeyp-72e1.7 |
| chr22_+_24389135 | 1.30 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr5_+_42957503 | 1.30 |
ENSDART00000192885
|
mob1ba
|
MOB kinase activator 1Ba |
| chr7_-_71837213 | 1.28 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr16_-_32208728 | 1.25 |
ENSDART00000023995
|
calhm5.2
|
calcium homeostasis modulator family member 5, tandem duplicate 2 |
| chr15_-_1484795 | 1.24 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr2_+_24199073 | 1.24 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr9_+_6587364 | 1.22 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr13_-_44782462 | 1.22 |
ENSDART00000141298
ENSDART00000099990 |
btbd9
|
BTB (POZ) domain containing 9 |
| chr1_+_46652615 | 1.20 |
ENSDART00000053221
|
arl11
|
ADP-ribosylation factor-like 11 |
| chr14_-_1990290 | 1.20 |
ENSDART00000183382
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr15_+_46329149 | 1.18 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr22_+_9918872 | 1.18 |
ENSDART00000177953
|
blf
|
bloody fingers |
| chr11_-_17803071 | 1.18 |
ENSDART00000080752
ENSDART00000153801 |
uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr16_+_46666074 | 1.17 |
ENSDART00000074919
ENSDART00000142698 |
ubqln4
|
ubiquilin 4 |
| chr9_+_6587056 | 1.17 |
ENSDART00000193421
|
fhl2a
|
four and a half LIM domains 2a |
| chr10_+_33573838 | 1.13 |
ENSDART00000051197
ENSDART00000130093 |
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr12_+_49135755 | 1.12 |
ENSDART00000153460
|
si:zfos-911d5.4
|
si:zfos-911d5.4 |
| chr20_-_27864964 | 1.10 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr6_-_39631164 | 1.08 |
ENSDART00000104042
ENSDART00000136076 |
atf7b
|
activating transcription factor 7b |
| chr9_+_6802641 | 1.00 |
ENSDART00000187278
|
FO203514.1
|
|
| chr2_-_48966431 | 0.98 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr18_+_50907675 | 0.96 |
ENSDART00000159950
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr21_+_27297145 | 0.96 |
ENSDART00000146588
|
si:dkey-175m17.6
|
si:dkey-175m17.6 |
| chr22_+_2229964 | 0.95 |
ENSDART00000112582
|
znf1161
|
zinc finger protein 1161 |
| chr8_+_13364950 | 0.94 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr10_-_11397590 | 0.94 |
ENSDART00000064212
|
srek1ip1
|
SREK1-interacting protein 1 |
| chr10_-_26273629 | 0.93 |
ENSDART00000147790
|
dchs1b
|
dachsous cadherin-related 1b |
| chr23_-_24047054 | 0.90 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr3_-_32290096 | 0.89 |
ENSDART00000148763
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr6_-_31323984 | 0.87 |
ENSDART00000183660
ENSDART00000184146 ENSDART00000180156 |
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr16_+_31827502 | 0.86 |
ENSDART00000045210
|
mlf2
|
myeloid leukemia factor 2 |
| chr1_-_53714885 | 0.86 |
ENSDART00000026409
|
cct4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr17_-_50018133 | 0.85 |
ENSDART00000112267
|
filip1a
|
filamin A interacting protein 1a |
| chr7_-_28463106 | 0.84 |
ENSDART00000137799
|
trim66
|
tripartite motif containing 66 |
| chr5_+_27267186 | 0.82 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr8_+_52637507 | 0.82 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr3_-_53722182 | 0.81 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr24_-_21808816 | 0.79 |
ENSDART00000025621
ENSDART00000130446 |
flt1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) |
| chr21_-_36948 | 0.79 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr21_-_13856689 | 0.79 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr15_+_18863875 | 0.78 |
ENSDART00000062603
|
cadm1b
|
cell adhesion molecule 1b |
| chr15_-_13254480 | 0.78 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr7_-_20464468 | 0.77 |
ENSDART00000134700
|
cnpy4
|
canopy4 |
| chr19_+_14158075 | 0.76 |
ENSDART00000170335
ENSDART00000168260 |
nudc
|
nudC nuclear distribution protein |
| chr2_-_6482240 | 0.76 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr6_-_29159888 | 0.75 |
ENSDART00000110288
|
zbtb11
|
zinc finger and BTB domain containing 11 |
| chr4_+_35553514 | 0.72 |
ENSDART00000182938
|
CR847906.1
|
|
| chr11_+_37144328 | 0.71 |
ENSDART00000162830
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr13_+_18202703 | 0.71 |
ENSDART00000109642
|
tet1
|
tet methylcytosine dioxygenase 1 |
| chr2_-_20866758 | 0.71 |
ENSDART00000165374
|
tpra
|
translocated promoter region a, nuclear basket protein |
| chr21_+_44581654 | 0.70 |
ENSDART00000187201
ENSDART00000180360 ENSDART00000191543 ENSDART00000180039 ENSDART00000186308 |
TMEM164
|
transmembrane protein 164 |
| chr5_-_23200880 | 0.68 |
ENSDART00000051531
|
iqcd
|
IQ motif containing D |
| chr1_+_53714734 | 0.67 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr6_+_49095646 | 0.67 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr18_+_8346920 | 0.61 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr10_-_26274094 | 0.60 |
ENSDART00000108798
|
dchs1b
|
dachsous cadherin-related 1b |
| chr5_-_22602979 | 0.59 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr2_-_9260002 | 0.58 |
ENSDART00000057299
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr23_+_24931999 | 0.58 |
ENSDART00000136162
ENSDART00000140335 |
klhl21
|
kelch-like family member 21 |
| chr18_+_3338228 | 0.54 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr1_+_10305611 | 0.53 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr19_+_23301050 | 0.52 |
ENSDART00000193713
|
si:dkey-79i2.4
|
si:dkey-79i2.4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-18a10.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 22.7 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 2.6 | 7.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 2.5 | 7.6 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 1.4 | 5.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 1.2 | 3.5 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.9 | 2.8 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.9 | 2.7 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.8 | 6.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.8 | 2.3 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.7 | 14.9 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.6 | 3.9 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.6 | 6.8 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.6 | 2.4 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.6 | 5.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.6 | 3.9 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.5 | 1.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.5 | 2.6 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.5 | 1.5 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.5 | 1.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 2.7 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.4 | 5.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.4 | 4.7 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.4 | 1.2 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.4 | 2.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.4 | 1.6 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.3 | 3.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 2.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 1.6 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.3 | 3.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 1.8 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.3 | 2.3 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.3 | 11.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 11.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 3.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 3.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.2 | 4.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 2.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 2.6 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.2 | 1.3 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.2 | 0.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 2.1 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 4.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 6.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.2 | 2.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.2 | 3.2 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 2.0 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.7 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 2.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 3.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.8 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 2.7 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.1 | 0.7 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 1.5 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 0.5 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.1 | 0.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 10.7 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.1 | 2.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.5 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 2.9 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 1.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.8 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.5 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.3 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.2 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.1 | 3.3 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 2.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 1.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.7 | GO:0043490 | C4-dicarboxylate transport(GO:0015740) acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.5 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.8 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 1.6 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.1 | 5.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 4.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 1.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 11.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.3 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.7 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 4.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 13.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 2.1 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.0 | 2.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.6 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 2.0 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 1.9 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 2.4 | GO:0001666 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.0 | 3.7 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 7.1 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 1.8 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 1.6 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 3.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 5.3 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.4 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 4.1 | GO:0008380 | RNA splicing(GO:0008380) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 32.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.4 | 6.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 5.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 3.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 2.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 2.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 5.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 21.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 2.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 2.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 2.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 3.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 5.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.9 | GO:0005686 | U2 snRNP(GO:0005686) U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.5 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 4.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 4.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.3 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 3.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 5.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 3.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 2.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 7.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 3.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 5.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 8.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.5 | GO:0005911 | cell-cell junction(GO:0005911) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.4 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.9 | 3.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.8 | 2.4 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.6 | 9.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 23.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.6 | 2.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 2.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.4 | 3.9 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.4 | 2.1 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.3 | 2.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 6.8 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 4.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.3 | 3.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 1.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.3 | 6.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 5.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 26.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.3 | 3.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 2.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 2.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 2.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 1.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.2 | 14.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 1.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 5.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.3 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.8 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 6.0 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 2.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 16.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 0.5 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.1 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 4.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 2.1 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 1.1 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 7.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.7 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.1 | 2.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 18.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 2.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 5.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 2.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 5.5 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.6 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 1.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 4.2 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 6.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 7.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 3.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 14.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 2.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.9 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 3.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 2.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 2.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 7.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 5.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 5.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |