Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
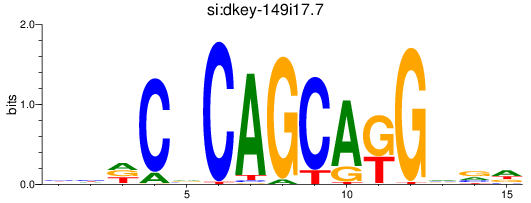
Results for si:dkey-149i17.7_zic1
Z-value: 1.19
Transcription factors associated with si:dkey-149i17.7_zic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-149i17.7
|
ENSDARG00000094684 | si_dkey-149i17.7 |
|
zic1
|
ENSDARG00000015567 | zic family member 1 (odd-paired homolog, Drosophila) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zic1 | dr11_v1_chr24_-_4973765_4973765 | 0.64 | 5.4e-12 | Click! |
| si:dkey-149i17.7 | dr11_v1_chr2_+_24885987_24885987 | 0.03 | 7.7e-01 | Click! |
Activity profile of si:dkey-149i17.7_zic1 motif
Sorted Z-values of si:dkey-149i17.7_zic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_24699985 | 12.53 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr23_-_46217134 | 10.56 |
ENSDART00000189477
ENSDART00000168352 |
CABZ01102528.1
|
|
| chr16_-_27640995 | 10.25 |
ENSDART00000019658
|
nacad
|
NAC alpha domain containing |
| chr14_-_51855047 | 10.18 |
ENSDART00000088912
|
cplx1
|
complexin 1 |
| chr22_+_5106751 | 9.70 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr13_+_27314795 | 9.40 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr24_-_3419998 | 9.28 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr3_+_24482999 | 9.04 |
ENSDART00000059179
|
nptxra
|
neuronal pentraxin receptor a |
| chr25_-_29134654 | 8.66 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr13_-_226393 | 8.62 |
ENSDART00000172677
|
rtn4b
|
reticulon 4b |
| chr8_+_8845932 | 8.57 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr18_+_49411417 | 8.34 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr18_+_7345417 | 8.01 |
ENSDART00000041429
|
glipr1b
|
GLI pathogenesis-related 1b |
| chr21_+_3093419 | 7.99 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr13_-_11667661 | 7.98 |
ENSDART00000102411
|
dctn1b
|
dynactin 1b |
| chr14_-_3031810 | 7.90 |
ENSDART00000090213
|
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr13_+_7292061 | 7.78 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr14_-_3032016 | 7.72 |
ENSDART00000183461
ENSDART00000183035 |
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr11_-_18799827 | 7.66 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr13_-_226109 | 7.63 |
ENSDART00000161705
ENSDART00000172744 ENSDART00000163902 ENSDART00000158208 |
rtn4b
|
reticulon 4b |
| chr17_+_5915875 | 7.28 |
ENSDART00000184179
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr16_-_55028740 | 7.10 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr24_-_30096666 | 7.04 |
ENSDART00000183285
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr1_+_45085194 | 7.01 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr19_+_46259619 | 6.99 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr10_-_2526526 | 6.95 |
ENSDART00000191726
ENSDART00000192767 |
CU856539.1
|
|
| chr18_+_5549672 | 6.39 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr14_+_7140997 | 6.28 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr6_-_13308813 | 6.25 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr4_-_2162688 | 6.06 |
ENSDART00000148900
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr20_-_18731268 | 6.00 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr12_-_35393211 | 5.79 |
ENSDART00000137139
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr19_-_32641725 | 5.64 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr7_-_67842997 | 5.56 |
ENSDART00000169763
|
pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr23_+_44741500 | 5.48 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr3_+_54047342 | 5.42 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr12_+_48634927 | 5.29 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr23_+_216012 | 5.27 |
ENSDART00000181115
ENSDART00000004678 ENSDART00000190439 ENSDART00000189322 |
PDZD4
|
si:ch73-162j3.4 |
| chr25_-_37205061 | 5.20 |
ENSDART00000155620
|
si:dkey-234i14.2
|
si:dkey-234i14.2 |
| chr25_-_10564721 | 5.20 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr4_-_1324141 | 5.10 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr5_-_45651548 | 5.10 |
ENSDART00000097645
|
NPFFR2
|
neuropeptide FF receptor 2a |
| chr19_-_9472893 | 5.09 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr25_+_31264155 | 5.08 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr9_+_307863 | 5.05 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr4_-_4795205 | 5.04 |
ENSDART00000039313
|
zgc:162331
|
zgc:162331 |
| chr16_-_13004166 | 5.01 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr4_-_26322834 | 4.96 |
ENSDART00000170709
ENSDART00000161687 |
st8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr17_+_13664442 | 4.75 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr16_+_14029283 | 4.60 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr9_-_6208067 | 4.58 |
ENSDART00000032368
|
tmtops2b
|
teleost multiple tissue opsin 2b |
| chr25_-_10565006 | 4.45 |
ENSDART00000130608
ENSDART00000190212 |
galn
|
galanin/GMAP prepropeptide |
| chr21_+_18353703 | 4.44 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr6_+_29923593 | 4.39 |
ENSDART00000169687
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr13_+_30912117 | 4.34 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr16_+_50741154 | 4.31 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr16_+_50755133 | 4.31 |
ENSDART00000029283
|
IGLON5
|
zgc:110372 |
| chr4_-_22671469 | 4.26 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr23_+_35759843 | 4.07 |
ENSDART00000047082
|
gdap1l1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr23_+_22267374 | 4.06 |
ENSDART00000079035
|
rap1gap
|
RAP1 GTPase activating protein |
| chr20_-_52846212 | 4.06 |
ENSDART00000040265
|
tbc1d7
|
TBC1 domain family, member 7 |
| chr21_-_45588720 | 3.96 |
ENSDART00000186642
ENSDART00000189531 |
LO018363.2
|
|
| chr10_-_41450367 | 3.95 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr3_-_19899914 | 3.89 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr17_-_44584811 | 3.89 |
ENSDART00000165059
ENSDART00000165252 |
slc35f4
|
solute carrier family 35, member F4 |
| chr7_+_22543963 | 3.85 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr14_+_36521553 | 3.78 |
ENSDART00000136233
|
TENM3
|
si:dkey-237h12.3 |
| chr25_+_27923846 | 3.76 |
ENSDART00000047007
|
slc13a1
|
solute carrier family 13 member 1 |
| chr22_+_15438513 | 3.59 |
ENSDART00000010846
|
gpc5b
|
glypican 5b |
| chr8_+_31777633 | 3.59 |
ENSDART00000142519
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr3_-_21061931 | 3.49 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr4_+_8376362 | 3.43 |
ENSDART00000138653
ENSDART00000132647 |
erc1b
|
ELKS/RAB6-interacting/CAST family member 1b |
| chr22_-_237651 | 3.40 |
ENSDART00000075210
|
zgc:66156
|
zgc:66156 |
| chr18_+_1615 | 3.40 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr18_+_3169579 | 3.39 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr13_-_41482064 | 3.34 |
ENSDART00000188322
ENSDART00000164732 |
pcdh15a
|
protocadherin-related 15a |
| chr8_+_28667236 | 3.31 |
ENSDART00000101159
|
slc13a3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr17_+_33296852 | 3.28 |
ENSDART00000154580
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr17_-_3303805 | 3.25 |
ENSDART00000169136
|
CABZ01007222.1
|
|
| chr18_+_3243292 | 3.22 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr1_+_23230459 | 3.14 |
ENSDART00000147857
|
si:dkey-92j12.5
|
si:dkey-92j12.5 |
| chr14_+_14225048 | 3.12 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr13_+_2523032 | 3.10 |
ENSDART00000172261
|
lhb
|
luteinizing hormone, beta polypeptide |
| chr9_+_55075526 | 3.05 |
ENSDART00000139555
ENSDART00000133864 |
gpr143
|
G protein-coupled receptor 143 |
| chr11_-_37411492 | 3.05 |
ENSDART00000166468
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr8_-_7386978 | 3.05 |
ENSDART00000058321
|
lhfpl4b
|
LHFPL tetraspan subfamily member 4b |
| chr3_+_14317802 | 3.01 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr10_-_22918214 | 3.00 |
ENSDART00000163908
|
rnasekb
|
ribonuclease, RNase K b |
| chr14_+_52413846 | 2.96 |
ENSDART00000160952
|
noa1
|
nitric oxide associated 1 |
| chr25_-_37435060 | 2.96 |
ENSDART00000102855
ENSDART00000148566 |
pdhx
|
pyruvate dehydrogenase complex, component X |
| chr2_+_3472832 | 2.95 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr13_+_30912385 | 2.89 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr6_+_10338554 | 2.89 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr15_+_47386939 | 2.86 |
ENSDART00000128224
|
FO904873.1
|
|
| chr8_+_54055390 | 2.86 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr14_-_36409960 | 2.85 |
ENSDART00000173140
|
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr24_-_28711176 | 2.85 |
ENSDART00000105753
|
olfm3a
|
olfactomedin 3a |
| chr10_+_7182423 | 2.81 |
ENSDART00000186788
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr5_-_72178739 | 2.76 |
ENSDART00000050971
|
rab14l
|
RAB14, member RAS oncogene family, like |
| chr2_-_15041846 | 2.75 |
ENSDART00000139050
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr8_+_28667021 | 2.73 |
ENSDART00000184292
|
slc13a3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr15_+_45591669 | 2.73 |
ENSDART00000157459
|
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr18_-_46010 | 2.72 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr4_-_4535189 | 2.70 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr11_-_12512122 | 2.67 |
ENSDART00000145338
|
si:dkey-27d5.3
|
si:dkey-27d5.3 |
| chr9_-_22158784 | 2.65 |
ENSDART00000167850
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr14_+_14224730 | 2.62 |
ENSDART00000180112
ENSDART00000184891 ENSDART00000174760 |
nlgn3a
|
neuroligin 3a |
| chr14_+_19258702 | 2.62 |
ENSDART00000187087
ENSDART00000005738 |
slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr12_-_4651988 | 2.60 |
ENSDART00000182836
|
si:ch211-255p10.4
|
si:ch211-255p10.4 |
| chr22_+_15438872 | 2.58 |
ENSDART00000139800
|
gpc5b
|
glypican 5b |
| chr17_-_6382392 | 2.55 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr21_-_4793686 | 2.52 |
ENSDART00000158232
|
notch1a
|
notch 1a |
| chr7_-_24994722 | 2.46 |
ENSDART00000131671
|
rcor2
|
REST corepressor 2 |
| chr17_-_20979077 | 2.46 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr1_-_625875 | 2.45 |
ENSDART00000167331
|
appa
|
amyloid beta (A4) precursor protein a |
| chr11_+_30057762 | 2.44 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr7_-_52842007 | 2.43 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr5_-_21044693 | 2.42 |
ENSDART00000140298
|
si:dkey-13n15.2
|
si:dkey-13n15.2 |
| chr23_-_28294763 | 2.39 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr9_+_34380299 | 2.37 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr7_+_36035432 | 2.37 |
ENSDART00000179004
|
irx3a
|
iroquois homeobox 3a |
| chr12_+_40905427 | 2.35 |
ENSDART00000170526
ENSDART00000185771 ENSDART00000193945 |
CDH18
|
cadherin 18 |
| chr13_+_31172833 | 2.35 |
ENSDART00000176378
|
CR931802.3
|
|
| chr17_-_52587598 | 2.34 |
ENSDART00000061497
|
si:ch211-173a9.6
|
si:ch211-173a9.6 |
| chr13_+_30912938 | 2.26 |
ENSDART00000190003
|
drgx
|
dorsal root ganglia homeobox |
| chr21_+_29077509 | 2.26 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr8_-_44015210 | 2.24 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr22_+_12366516 | 2.23 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr6_-_44711942 | 2.21 |
ENSDART00000055035
|
cntn3b
|
contactin 3b |
| chr6_+_518979 | 2.20 |
ENSDART00000151012
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr4_+_38344 | 2.15 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr15_-_16323750 | 2.13 |
ENSDART00000028500
|
nxn
|
nucleoredoxin |
| chr6_-_3978919 | 2.12 |
ENSDART00000167753
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr13_-_8279533 | 2.12 |
ENSDART00000109666
|
si:ch211-250c4.5
|
si:ch211-250c4.5 |
| chr2_+_28889936 | 2.08 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr15_+_24691088 | 2.08 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr21_-_40557281 | 2.07 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr2_-_53896300 | 2.06 |
ENSDART00000161221
|
capsla
|
calcyphosine-like a |
| chr16_-_20492799 | 2.04 |
ENSDART00000014769
|
chn2
|
chimerin 2 |
| chr6_+_42587637 | 2.01 |
ENSDART00000179964
|
camkva
|
CaM kinase-like vesicle-associated a |
| chr14_-_4043818 | 2.01 |
ENSDART00000179870
|
snx25
|
sorting nexin 25 |
| chr1_+_40562540 | 2.00 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr13_-_48756036 | 2.00 |
ENSDART00000035018
ENSDART00000132895 |
ntpcr
|
nucleoside-triphosphatase, cancer-related |
| chr16_+_31511739 | 1.99 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr12_+_31735159 | 1.97 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr24_+_12200260 | 1.95 |
ENSDART00000181391
|
CR855257.2
|
|
| chr7_+_58843700 | 1.94 |
ENSDART00000159500
ENSDART00000158436 |
lypla1
|
lysophospholipase I |
| chr12_-_6551681 | 1.89 |
ENSDART00000145413
|
si:ch211-253p2.2
|
si:ch211-253p2.2 |
| chr18_+_50278858 | 1.87 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr7_-_24994568 | 1.87 |
ENSDART00000002961
|
rcor2
|
REST corepressor 2 |
| chr1_-_21529641 | 1.80 |
ENSDART00000135905
|
wdr19
|
WD repeat domain 19 |
| chr22_-_20924564 | 1.78 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr13_-_25632756 | 1.77 |
ENSDART00000077627
ENSDART00000139237 |
ret
|
ret proto-oncogene receptor tyrosine kinase |
| chr17_-_14836320 | 1.75 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr4_+_5798223 | 1.73 |
ENSDART00000059440
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr20_+_1960092 | 1.70 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr20_+_26892761 | 1.68 |
ENSDART00000133293
|
ftr97
|
finTRIM family, member 97 |
| chr20_+_11800063 | 1.67 |
ENSDART00000152230
|
si:ch211-155o21.4
|
si:ch211-155o21.4 |
| chr10_+_8680730 | 1.65 |
ENSDART00000011987
|
isl1l
|
islet1, like |
| chr19_+_1999838 | 1.64 |
ENSDART00000166669
|
btd
|
biotinidase |
| chr1_-_1627487 | 1.63 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr23_+_16620801 | 1.61 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr22_+_2207502 | 1.61 |
ENSDART00000169162
|
si:dkeyp-79b7.12
|
si:dkeyp-79b7.12 |
| chr24_+_42948 | 1.60 |
ENSDART00000122785
|
tmx3b
|
thioredoxin related transmembrane protein 3b |
| chr22_-_3449282 | 1.60 |
ENSDART00000136798
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr11_+_30058139 | 1.60 |
ENSDART00000112254
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr8_+_26874924 | 1.55 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr15_+_1119114 | 1.55 |
ENSDART00000102224
|
p2ry12
|
purinergic receptor P2Y12 |
| chr25_+_26923193 | 1.54 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr23_-_28692581 | 1.54 |
ENSDART00000078141
|
ribc1
|
RIB43A domain with coiled-coils 1 |
| chr2_-_10632431 | 1.54 |
ENSDART00000122709
|
mtf2
|
metal response element binding transcription factor 2 |
| chr2_+_25019387 | 1.54 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr1_+_41690402 | 1.53 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr4_-_10958074 | 1.53 |
ENSDART00000150478
|
si:ch211-161n3.4
|
si:ch211-161n3.4 |
| chr16_+_25296389 | 1.52 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr14_+_8328645 | 1.51 |
ENSDART00000127494
|
nrg2b
|
neuregulin 2b |
| chr12_+_47909026 | 1.49 |
ENSDART00000192472
|
tbata
|
thymus, brain and testes associated |
| chr15_-_25613114 | 1.49 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr23_+_21414005 | 1.48 |
ENSDART00000144686
|
iffo2a
|
intermediate filament family orphan 2a |
| chr10_+_16092671 | 1.46 |
ENSDART00000182761
ENSDART00000154835 |
megf10
|
multiple EGF-like-domains 10 |
| chr17_-_52941606 | 1.44 |
ENSDART00000155310
|
zgc:154061
|
zgc:154061 |
| chr7_-_36358735 | 1.43 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr7_-_36358303 | 1.43 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr18_-_45736 | 1.40 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr8_-_50346246 | 1.39 |
ENSDART00000025008
|
stc1
|
stanniocalcin 1 |
| chr13_-_133520 | 1.38 |
ENSDART00000180874
|
FO904997.1
|
|
| chr7_-_59514547 | 1.37 |
ENSDART00000168457
|
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr10_+_33990572 | 1.37 |
ENSDART00000138547
|
fryb
|
furry homolog b (Drosophila) |
| chr23_+_2361184 | 1.36 |
ENSDART00000184469
|
CABZ01048666.1
|
|
| chr11_-_45141309 | 1.35 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr14_-_38843690 | 1.35 |
ENSDART00000183629
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr5_-_68779747 | 1.35 |
ENSDART00000192636
ENSDART00000188039 |
mepce
|
methylphosphate capping enzyme |
| chr6_-_32726848 | 1.34 |
ENSDART00000155294
|
zc3h3
|
zinc finger CCCH-type containing 3 |
| chr3_+_14339728 | 1.34 |
ENSDART00000184127
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr21_-_13225402 | 1.33 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr7_-_5396154 | 1.32 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr3_-_18575868 | 1.31 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr21_-_22709251 | 1.29 |
ENSDART00000140032
|
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr8_+_7756893 | 1.29 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr15_+_34062460 | 1.28 |
ENSDART00000164654
|
si:dkey-30e9.6
|
si:dkey-30e9.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-149i17.7_zic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.2 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 3.1 | 12.5 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 1.6 | 6.4 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.4 | 15.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 1.4 | 4.1 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 1.1 | 6.6 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 1.0 | 5.0 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 1.0 | 2.9 | GO:0042245 | RNA repair(GO:0042245) |
| 0.9 | 9.7 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.8 | 2.5 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.7 | 3.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.7 | 2.8 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.6 | 7.0 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.6 | 6.0 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.6 | 13.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.5 | 2.7 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.5 | 5.7 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.5 | 7.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 1.3 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 3.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.4 | 3.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.4 | 4.3 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.4 | 3.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.4 | 1.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.4 | 1.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.4 | 1.5 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.4 | 1.8 | GO:1901166 | branchiomeric skeletal muscle development(GO:0014707) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.3 | 5.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 9.4 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.3 | 1.6 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.3 | 9.3 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.3 | 0.9 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.3 | 1.8 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 5.5 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.3 | 2.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.3 | 2.0 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 4.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 0.7 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 8.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 5.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.7 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.2 | 2.1 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 3.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 2.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.2 | 1.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.2 | 1.0 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 1.2 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 1.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 1.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 2.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.9 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 5.8 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 0.7 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 3.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 11.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 1.2 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.4 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 1.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.6 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 10.3 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.1 | 0.4 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 1.6 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 2.1 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.4 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.1 | 6.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.9 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.4 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.3 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 2.0 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 2.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 4.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 4.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 5.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 0.7 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.2 | GO:0048327 | axial mesoderm formation(GO:0048320) axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 1.1 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 3.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 1.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 2.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 6.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 3.1 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 6.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 2.1 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 1.3 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 3.0 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.4 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 1.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 4.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 3.6 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 1.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.5 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 3.2 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 2.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 2.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 2.6 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.7 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 1.5 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.4 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 1.4 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.8 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.5 | GO:0030239 | myofibril assembly(GO:0030239) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.1 | 5.6 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.5 | 5.5 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.5 | 12.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 3.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.3 | 6.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 3.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 5.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.3 | 8.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 6.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 3.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.4 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 1.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 6.5 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.2 | 4.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 0.5 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 9.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 2.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 4.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 4.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.4 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 2.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.5 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 5.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 2.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.0 | 1.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 11.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 1.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 2.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 16.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 8.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 6.2 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 8.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 15.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.6 | 6.4 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 1.5 | 6.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 1.1 | 4.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.0 | 3.1 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.9 | 9.7 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.7 | 3.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.7 | 2.9 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.7 | 7.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.7 | 6.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 5.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 7.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.5 | 2.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.5 | 1.5 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.4 | 11.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.4 | 3.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.4 | 6.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.3 | 2.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.3 | 1.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.3 | 5.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 2.7 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.2 | 1.6 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.2 | 8.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 1.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 0.6 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 2.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 4.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 3.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 6.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 8.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 7.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 4.1 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 6.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 2.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 6.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 5.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.9 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 10.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 2.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.4 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 1.5 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 4.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 5.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 1.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 4.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 4.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 2.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 27.8 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 3.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 3.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 1.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 7.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.3 | 5.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 1.8 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 4.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 8.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 7.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.6 | 9.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 8.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.5 | 4.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 3.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 4.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 5.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 3.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 6.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.9 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 6.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 4.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 4.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |