Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
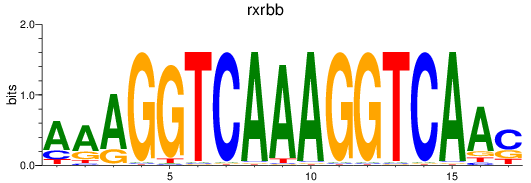
Results for rxrbb
Z-value: 0.72
Transcription factors associated with rxrbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rxrbb
|
ENSDARG00000002006 | retinoid x receptor, beta b |
|
rxrbb
|
ENSDARG00000117079 | retinoid x receptor, beta b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rxrbb | dr11_v1_chr16_+_18535618_18535618 | 0.63 | 7.4e-12 | Click! |
Activity profile of rxrbb motif
Sorted Z-values of rxrbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_25848231 | 8.13 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr13_-_9318891 | 7.98 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr23_-_45504991 | 7.95 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr3_+_30257582 | 6.64 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr25_+_21829777 | 6.57 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr18_-_48517040 | 6.42 |
ENSDART00000143645
|
kcnj1a.3
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 3 |
| chr7_-_38633867 | 6.09 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr10_+_45128375 | 5.95 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr10_-_17745345 | 5.34 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr18_-_48492951 | 5.28 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr18_-_48508585 | 5.19 |
ENSDART00000133364
|
kcnj1a.4
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 4 |
| chr10_+_20128267 | 4.61 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr8_+_36803415 | 4.56 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr13_+_28819768 | 3.92 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr2_+_42191592 | 3.88 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr25_+_31238606 | 3.77 |
ENSDART00000149439
|
tnni2a.2
|
troponin I type 2a (skeletal, fast), tandem duplicate 2 |
| chr21_-_26495700 | 3.68 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr24_-_21172122 | 3.66 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr5_-_23362602 | 3.42 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr13_+_28821841 | 3.21 |
ENSDART00000179900
|
CU639469.1
|
|
| chr25_-_19420949 | 3.15 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr6_-_11523987 | 3.11 |
ENSDART00000189363
|
gulp1b
|
GULP, engulfment adaptor PTB domain containing 1b |
| chr20_-_53078607 | 2.97 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr23_+_30730121 | 2.97 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr11_-_11910225 | 2.75 |
ENSDART00000159922
|
si:ch211-69b7.6
|
si:ch211-69b7.6 |
| chr1_+_23783349 | 2.63 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr23_-_42810664 | 2.59 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr19_+_3056450 | 2.58 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr5_+_38276582 | 2.57 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr18_+_27515640 | 2.54 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr22_-_3595439 | 2.53 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr1_+_58353661 | 2.52 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr18_-_48550426 | 2.49 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr9_+_54644626 | 2.45 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr19_+_2275019 | 2.42 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr6_+_40775800 | 2.41 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr19_+_2631565 | 2.40 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr2_-_22659450 | 2.38 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr5_-_27994679 | 2.33 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr17_+_11675362 | 2.30 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr18_+_31410652 | 2.27 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr24_-_1021318 | 2.25 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr8_+_1009831 | 2.19 |
ENSDART00000172414
|
fabp1b.2
|
fatty acid binding protein 1b, liver, tandem duplicate 2 |
| chr19_+_177455 | 2.15 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr8_-_979735 | 2.11 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr15_+_47618221 | 2.06 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr15_+_47386939 | 2.06 |
ENSDART00000128224
|
FO904873.1
|
|
| chr6_+_60036767 | 2.04 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr1_+_58182400 | 2.01 |
ENSDART00000144784
|
si:ch211-15j1.1
|
si:ch211-15j1.1 |
| chr1_+_58119117 | 2.01 |
ENSDART00000146788
|
si:ch211-15j1.5
|
si:ch211-15j1.5 |
| chr5_-_33255759 | 2.00 |
ENSDART00000085531
|
prkaa1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr25_+_20119466 | 1.89 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr2_-_32555625 | 1.87 |
ENSDART00000056641
ENSDART00000137531 |
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr25_-_31118923 | 1.86 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr24_-_29586082 | 1.85 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr15_+_15856178 | 1.76 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr13_-_4018888 | 1.72 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr16_+_14033121 | 1.69 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr1_+_58303892 | 1.69 |
ENSDART00000147678
|
CR769768.1
|
|
| chr23_+_41679586 | 1.67 |
ENSDART00000067662
|
CU914487.1
|
|
| chr22_-_28668442 | 1.67 |
ENSDART00000182377
|
col8a1b
|
collagen, type VIII, alpha 1b |
| chr2_-_8102169 | 1.66 |
ENSDART00000131955
|
pls1
|
plastin 1 (I isoform) |
| chr9_-_37613792 | 1.65 |
ENSDART00000138345
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr9_+_34380299 | 1.59 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr12_-_31012741 | 1.54 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7 like 2 |
| chr9_+_22782027 | 1.53 |
ENSDART00000090816
|
rif1
|
replication timing regulatory factor 1 |
| chr1_+_58442694 | 1.53 |
ENSDART00000160897
|
zgc:194906
|
zgc:194906 |
| chr1_+_58150000 | 1.51 |
ENSDART00000137836
|
si:ch211-15j1.3
|
si:ch211-15j1.3 |
| chr17_-_7371564 | 1.48 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr23_-_18595020 | 1.46 |
ENSDART00000187032
ENSDART00000191047 |
ikbkg
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr1_+_58139102 | 1.44 |
ENSDART00000134826
|
si:ch211-15j1.4
|
si:ch211-15j1.4 |
| chr6_+_39930825 | 1.44 |
ENSDART00000182352
ENSDART00000186643 ENSDART00000190513 ENSDART00000065092 ENSDART00000188645 |
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr20_-_51656512 | 1.44 |
ENSDART00000129965
|
LO018154.1
|
|
| chr17_+_45737992 | 1.44 |
ENSDART00000135073
ENSDART00000143525 ENSDART00000158165 ENSDART00000184167 ENSDART00000109525 |
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr4_+_68519945 | 1.39 |
ENSDART00000171148
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr16_+_25809235 | 1.38 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr21_+_1119046 | 1.35 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr1_+_58260886 | 1.30 |
ENSDART00000110453
|
si:dkey-222h21.8
|
si:dkey-222h21.8 |
| chr24_-_1341543 | 1.27 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr19_-_3056235 | 1.27 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr12_+_46708920 | 1.26 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr4_-_38421217 | 1.26 |
ENSDART00000164517
|
znf1092
|
szinc finger protein 1092 |
| chr17_-_20711735 | 1.20 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr23_-_27050083 | 1.19 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr18_+_30441740 | 1.17 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr1_-_58868306 | 1.16 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr19_-_9867001 | 1.14 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr3_+_12755535 | 1.13 |
ENSDART00000161286
|
cyp2k17
|
cytochrome P450, family 2, subfamily K, polypeptide17 |
| chr4_-_16451375 | 1.12 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr16_-_29146624 | 1.07 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr9_+_32978302 | 1.07 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr21_-_35832548 | 1.01 |
ENSDART00000180840
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr10_+_589501 | 1.00 |
ENSDART00000188415
|
LO018557.1
|
|
| chr20_-_18382708 | 1.00 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr1_+_58312187 | 0.99 |
ENSDART00000142285
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr19_+_11214007 | 0.95 |
ENSDART00000127362
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr17_+_21817382 | 0.94 |
ENSDART00000079011
ENSDART00000189387 |
ikzf5
|
IKAROS family zinc finger 5 |
| chr4_-_858434 | 0.94 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr4_-_37587110 | 0.93 |
ENSDART00000169302
|
znf1100
|
zinc finger protein 1100 |
| chr16_-_24612871 | 0.92 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr15_+_22448456 | 0.91 |
ENSDART00000040542
ENSDART00000190270 ENSDART00000185897 |
arhgef12a
|
Rho guanine nucleotide exchange factor (GEF) 12a |
| chr2_+_36112273 | 0.91 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr19_+_5146460 | 0.90 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr21_-_43992027 | 0.90 |
ENSDART00000188612
|
cdx1b
|
caudal type homeobox 1 b |
| chr16_-_47483142 | 0.90 |
ENSDART00000147072
|
cthrc1b
|
collagen triple helix repeat containing 1b |
| chr17_-_27200634 | 0.88 |
ENSDART00000185332
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr13_+_45980163 | 0.88 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr14_-_13048355 | 0.85 |
ENSDART00000166434
|
si:dkey-35h6.1
|
si:dkey-35h6.1 |
| chr19_+_20713424 | 0.84 |
ENSDART00000129730
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr23_+_2914577 | 0.82 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr4_+_2655358 | 0.80 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr12_+_3848109 | 0.78 |
ENSDART00000144067
ENSDART00000192042 |
cabp5a
|
calcium binding protein 5a |
| chr2_-_22660232 | 0.76 |
ENSDART00000174742
|
thap4
|
THAP domain containing 4 |
| chr4_+_45274792 | 0.76 |
ENSDART00000150295
|
znf1138
|
zinc finger protein 1138 |
| chr7_-_8014186 | 0.74 |
ENSDART00000190012
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr7_-_8156169 | 0.73 |
ENSDART00000182960
ENSDART00000190184 ENSDART00000190885 ENSDART00000189867 ENSDART00000160836 |
si:cabz01030277.1
si:ch211-163c2.3
|
si:cabz01030277.1 si:ch211-163c2.3 |
| chr17_+_21817859 | 0.73 |
ENSDART00000143832
ENSDART00000141462 |
ikzf5
|
IKAROS family zinc finger 5 |
| chr20_-_26937453 | 0.67 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr1_+_58312680 | 0.66 |
ENSDART00000138007
|
si:dkey-222h21.10
|
si:dkey-222h21.10 |
| chr24_+_39034090 | 0.64 |
ENSDART00000185763
|
capn15
|
calpain 15 |
| chr18_+_27926839 | 0.60 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr2_-_7696503 | 0.59 |
ENSDART00000169709
|
CABZ01055306.1
|
|
| chr21_-_40015530 | 0.55 |
ENSDART00000084237
|
slc47a2.1
|
solute carrier family 47 (multidrug and toxin extrusion), member 2.1 |
| chr14_+_30413312 | 0.55 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr19_+_19032141 | 0.54 |
ENSDART00000192432
|
cpne4b
|
copine IVb |
| chr13_-_41546779 | 0.53 |
ENSDART00000163331
|
pcdh15a
|
protocadherin-related 15a |
| chr1_+_57050899 | 0.52 |
ENSDART00000152601
|
si:ch211-1f22.14
|
si:ch211-1f22.14 |
| chr15_+_45640906 | 0.46 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr5_-_20491198 | 0.46 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr13_+_21826369 | 0.43 |
ENSDART00000165150
ENSDART00000192115 |
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr5_-_32890807 | 0.43 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr9_-_25255490 | 0.41 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr25_+_15933411 | 0.39 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr24_+_81527 | 0.38 |
ENSDART00000192139
|
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr17_-_25395395 | 0.37 |
ENSDART00000170233
|
fam167b
|
family with sequence similarity 167, member B |
| chr18_-_158541 | 0.36 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr2_+_57801960 | 0.35 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr10_-_29892486 | 0.32 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr21_-_44512893 | 0.32 |
ENSDART00000166853
|
zgc:136410
|
zgc:136410 |
| chr20_-_26936887 | 0.31 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr13_-_36418921 | 0.31 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr12_+_17439580 | 0.27 |
ENSDART00000189257
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr7_+_25000060 | 0.24 |
ENSDART00000039265
ENSDART00000141814 |
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr12_-_33568174 | 0.18 |
ENSDART00000142329
|
tdrkh
|
tudor and KH domain containing |
| chr4_-_75172216 | 0.17 |
ENSDART00000127522
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr12_+_19958845 | 0.17 |
ENSDART00000193248
|
ercc4
|
excision repair cross-complementation group 4 |
| chr3_+_39566999 | 0.15 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr21_-_11820379 | 0.15 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr8_+_999421 | 0.14 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr7_+_7495034 | 0.05 |
ENSDART00000102629
ENSDART00000180475 |
nit1
|
nitrilase 1 |
| chr8_+_8012570 | 0.05 |
ENSDART00000183429
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr20_+_27519429 | 0.04 |
ENSDART00000136251
|
kif26aa
|
kinesin family member 26Aa |
| chr6_+_40952031 | 0.03 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr11_-_42230491 | 0.02 |
ENSDART00000164423
|
CABZ01030862.1
|
|
| chr6_-_14004772 | 0.01 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rxrbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.9 | 2.6 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.7 | 2.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.5 | 6.6 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 1.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.3 | 1.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.3 | 0.8 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.3 | 1.3 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.2 | 19.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.9 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.2 | 4.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 2.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.2 | 0.8 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.2 | 1.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 2.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 5.9 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 2.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.4 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 3.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 1.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.4 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 4.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.6 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 1.0 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 2.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 1.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 2.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 2.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 3.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 2.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.1 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.3 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 1.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.2 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 3.7 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 3.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 2.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 2.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.5 | GO:0032200 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 4.1 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 1.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 2.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.5 | 2.7 | GO:0043034 | costamere(GO:0043034) |
| 0.5 | 2.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 3.0 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.4 | 1.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.4 | 2.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 1.5 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 3.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 0.8 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 1.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 4.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 4.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 3.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 7.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 2.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.5 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 5.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.0 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 19.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.2 | 3.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 10.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.6 | 3.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.5 | 6.6 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 2.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 2.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 1.9 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.3 | 3.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 3.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 2.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.4 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.2 | 2.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 2.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 5.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 4.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.4 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.1 | 2.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.8 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 4.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 9.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 2.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.3 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 4.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.1 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 7.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 6.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.0 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 1.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.2 | 2.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 1.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 7.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 2.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 2.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |