Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
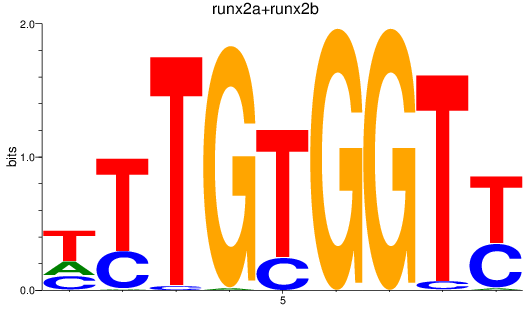
Results for runx2a+runx2b
Z-value: 1.43
Transcription factors associated with runx2a+runx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx2a
|
ENSDARG00000040261 | RUNX family transcription factor 2a |
|
runx2b
|
ENSDARG00000059233 | RUNX family transcription factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx2b | dr11_v1_chr20_-_44055095_44055123 | 0.67 | 2.1e-13 | Click! |
| runx2a | dr11_v1_chr17_+_5351922_5351922 | 0.18 | 8.3e-02 | Click! |
Activity profile of runx2a+runx2b motif
Sorted Z-values of runx2a+runx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_11334733 | 20.62 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr16_+_11724230 | 17.28 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr5_-_23277939 | 17.04 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr10_-_15128771 | 15.95 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr7_+_25858380 | 14.80 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr19_+_30633453 | 14.07 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr2_-_44282796 | 12.74 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr24_-_4973765 | 12.06 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr5_-_23277385 | 11.76 |
ENSDART00000134982
|
plp1b
|
proteolipid protein 1b |
| chr20_-_14054083 | 11.13 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr23_-_15878879 | 11.00 |
ENSDART00000010119
|
eef1a2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr2_-_44283554 | 10.83 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr2_-_44280061 | 10.60 |
ENSDART00000136818
|
mpz
|
myelin protein zero |
| chr13_+_27316934 | 10.25 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr13_+_7292061 | 9.99 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr13_+_27316632 | 9.78 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr3_+_28576173 | 9.78 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr15_+_29088426 | 8.35 |
ENSDART00000187290
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr3_-_22212764 | 7.82 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr25_+_4751879 | 7.70 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr22_-_33679277 | 7.57 |
ENSDART00000169948
|
FO904977.1
|
|
| chr9_+_42066030 | 7.45 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr25_+_34984333 | 7.34 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr9_-_40765868 | 7.27 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr7_-_12065668 | 7.25 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr20_-_30035326 | 7.16 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr16_+_31853919 | 7.14 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr15_+_6117502 | 7.08 |
ENSDART00000188293
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr1_+_15137901 | 7.07 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr14_+_38786298 | 6.86 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr7_-_2039060 | 6.82 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr23_+_27789795 | 6.74 |
ENSDART00000141458
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr8_+_36803415 | 6.73 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr4_-_2525916 | 6.63 |
ENSDART00000134123
ENSDART00000132581 ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr5_-_32445835 | 6.61 |
ENSDART00000170919
|
ncs1a
|
neuronal calcium sensor 1a |
| chr14_+_34514336 | 6.60 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr7_-_38633867 | 6.41 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr15_-_24869826 | 6.26 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr19_+_37857936 | 6.26 |
ENSDART00000189289
|
nxph1
|
neurexophilin 1 |
| chr11_-_27057572 | 6.22 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr17_+_19630272 | 6.15 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr23_-_27822920 | 6.08 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr13_-_10261383 | 5.98 |
ENSDART00000080808
|
six3a
|
SIX homeobox 3a |
| chr2_+_36898982 | 5.97 |
ENSDART00000084859
|
rabgap1l2
|
RAB GTPase activating protein 1-like 2 |
| chr7_+_46368520 | 5.97 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr1_-_8428736 | 5.96 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr16_+_50741154 | 5.80 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr21_-_35324091 | 5.79 |
ENSDART00000185042
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr2_-_22286828 | 5.75 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr7_-_33829824 | 5.70 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr3_+_37197686 | 5.70 |
ENSDART00000151144
|
fmnl1a
|
formin-like 1a |
| chr6_-_12314475 | 5.69 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr11_+_24703108 | 5.62 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr23_-_29376859 | 5.59 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr2_-_40135942 | 5.55 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr1_+_49266886 | 5.52 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr4_+_11375894 | 5.47 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr21_-_26495700 | 5.43 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr16_-_32672883 | 5.41 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr23_-_36724575 | 5.29 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr6_+_13806466 | 5.28 |
ENSDART00000043522
|
tmem198b
|
transmembrane protein 198b |
| chr18_-_41375120 | 5.20 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr5_-_52277643 | 5.01 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr16_+_32995882 | 4.99 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr8_-_17067364 | 4.98 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr21_+_25068215 | 4.95 |
ENSDART00000167523
ENSDART00000189259 |
dixdc1b
|
DIX domain containing 1b |
| chr7_+_38770167 | 4.95 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr5_+_23597144 | 4.94 |
ENSDART00000143929
ENSDART00000142420 ENSDART00000032909 |
kat5b
|
K(lysine) acetyltransferase 5b |
| chr6_+_23810529 | 4.85 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr10_+_21797276 | 4.84 |
ENSDART00000169105
|
pcdh1g29
|
protocadherin 1 gamma 29 |
| chr19_+_5146460 | 4.83 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr2_-_31936966 | 4.80 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr22_-_10459880 | 4.78 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr14_-_46616487 | 4.71 |
ENSDART00000105417
ENSDART00000166550 ENSDART00000105418 |
prom1a
|
prominin 1a |
| chr16_-_9869056 | 4.70 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr11_+_24994705 | 4.70 |
ENSDART00000129211
|
zgc:92107
|
zgc:92107 |
| chr22_+_9871238 | 4.69 |
ENSDART00000141085
ENSDART00000105939 |
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr19_+_40350468 | 4.67 |
ENSDART00000087444
|
hepacam2
|
HEPACAM family member 2 |
| chr4_-_8882572 | 4.67 |
ENSDART00000190060
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr1_-_26702930 | 4.65 |
ENSDART00000109297
ENSDART00000152389 |
foxe1
|
forkhead box E1 |
| chr6_+_38381957 | 4.61 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr21_+_13387965 | 4.61 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr10_-_17103651 | 4.61 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr13_-_29424454 | 4.57 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr9_+_35860975 | 4.55 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr4_+_17336557 | 4.55 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr21_+_15824182 | 4.51 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr7_-_40993456 | 4.49 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr5_-_6377865 | 4.47 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr7_+_31130667 | 4.44 |
ENSDART00000173937
|
tjp1a
|
tight junction protein 1a |
| chr25_-_35996141 | 4.41 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr7_-_18656069 | 4.37 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr19_+_16222618 | 4.36 |
ENSDART00000137189
ENSDART00000169246 ENSDART00000190583 ENSDART00000189521 |
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr13_+_25433774 | 4.31 |
ENSDART00000141255
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr19_-_9867001 | 4.29 |
ENSDART00000091695
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr25_+_21324588 | 4.27 |
ENSDART00000151842
|
lrrn3a
|
leucine rich repeat neuronal 3a |
| chr5_+_13373593 | 4.27 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr18_-_40508528 | 4.25 |
ENSDART00000185249
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr7_+_31051603 | 4.24 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr1_-_58064738 | 4.21 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr13_+_11440389 | 4.19 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr3_+_49021079 | 4.19 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr3_+_54168007 | 4.18 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr16_-_22863348 | 4.16 |
ENSDART00000147609
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr3_+_36160979 | 4.14 |
ENSDART00000038525
|
si:ch211-234h8.7
|
si:ch211-234h8.7 |
| chr13_-_23095006 | 4.13 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
| chr11_+_25139495 | 4.12 |
ENSDART00000168368
|
si:ch211-25d12.7
|
si:ch211-25d12.7 |
| chr10_-_7974155 | 4.12 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr20_+_5564042 | 4.11 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr3_+_30922947 | 4.08 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr22_+_21255860 | 4.06 |
ENSDART00000134893
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr15_-_12545683 | 4.00 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr6_+_29860776 | 3.99 |
ENSDART00000028406
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr12_+_32159272 | 3.98 |
ENSDART00000153167
|
hlfb
|
hepatic leukemia factor b |
| chr21_+_21309164 | 3.97 |
ENSDART00000132174
|
si:ch211-191j22.8
|
si:ch211-191j22.8 |
| chr2_+_12255568 | 3.93 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr8_+_21353878 | 3.92 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr2_+_22694382 | 3.90 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr10_+_17088261 | 3.90 |
ENSDART00000132103
|
si:dkey-106l3.7
|
si:dkey-106l3.7 |
| chr3_+_15271943 | 3.88 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr7_+_63325819 | 3.87 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr15_-_34214440 | 3.82 |
ENSDART00000167052
|
etv1
|
ets variant 1 |
| chr2_-_50298337 | 3.82 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr1_+_41854298 | 3.82 |
ENSDART00000192672
|
smox
|
spermine oxidase |
| chr17_-_33714636 | 3.81 |
ENSDART00000188500
|
DNAL1
|
si:dkey-84k17.3 |
| chr5_-_28606916 | 3.79 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr14_-_14746767 | 3.79 |
ENSDART00000183755
ENSDART00000190938 |
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr21_-_13672195 | 3.78 |
ENSDART00000146717
|
clic3
|
chloride intracellular channel 3 |
| chr18_+_35742838 | 3.78 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr16_+_25809235 | 3.74 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr24_-_15131831 | 3.68 |
ENSDART00000028410
|
cd226
|
CD226 molecule |
| chr5_+_62374092 | 3.68 |
ENSDART00000082965
|
CU928117.1
|
|
| chr11_-_30630628 | 3.67 |
ENSDART00000103265
|
zgc:158773
|
zgc:158773 |
| chr2_+_24722771 | 3.61 |
ENSDART00000147207
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr25_+_19954576 | 3.58 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr21_-_42100471 | 3.55 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr19_-_3482138 | 3.54 |
ENSDART00000168139
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr19_-_37154436 | 3.53 |
ENSDART00000103155
|
cx39.4
|
connexin 39.4 |
| chr15_+_39977461 | 3.53 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr9_-_37749973 | 3.53 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr15_+_25489406 | 3.51 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr23_-_7799184 | 3.51 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr22_+_20195280 | 3.51 |
ENSDART00000088603
ENSDART00000135692 |
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr2_-_16159491 | 3.50 |
ENSDART00000110059
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr2_+_31948352 | 3.48 |
ENSDART00000192611
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr21_+_21201346 | 3.47 |
ENSDART00000142961
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr13_+_40019001 | 3.44 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr20_-_35246150 | 3.42 |
ENSDART00000090549
|
fzd3a
|
frizzled class receptor 3a |
| chr11_-_42918971 | 3.40 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr19_+_42693855 | 3.40 |
ENSDART00000136873
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr18_+_17959681 | 3.31 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr13_+_17468161 | 3.31 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr3_-_36440705 | 3.31 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr4_+_20255160 | 3.26 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr8_-_39859688 | 3.26 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr8_+_25359394 | 3.25 |
ENSDART00000111248
|
si:dkey-183p4.10
|
si:dkey-183p4.10 |
| chr8_+_14778292 | 3.23 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr10_-_20453995 | 3.23 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr10_-_25769334 | 3.19 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr23_+_36653376 | 3.18 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr15_+_19838458 | 3.18 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr14_-_14566417 | 3.17 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr23_+_28582865 | 3.13 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr3_+_50310684 | 3.13 |
ENSDART00000112152
|
gas7a
|
growth arrest-specific 7a |
| chr25_+_20119466 | 3.12 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr7_+_65876335 | 3.11 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr16_+_29043813 | 3.11 |
ENSDART00000122681
|
nes
|
nestin |
| chr17_+_13031497 | 3.10 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr25_+_14109626 | 3.09 |
ENSDART00000109883
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr6_-_16394528 | 3.07 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr1_-_30689004 | 3.07 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr18_+_18612388 | 3.06 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr1_+_54650048 | 3.06 |
ENSDART00000141207
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr12_+_20336070 | 3.04 |
ENSDART00000066385
|
zgc:163057
|
zgc:163057 |
| chr6_-_24103666 | 3.04 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr17_-_46457622 | 3.01 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr20_+_38285671 | 2.98 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr21_+_34992550 | 2.97 |
ENSDART00000109041
ENSDART00000135400 |
tmprss15
|
transmembrane protease, serine 15 |
| chr6_-_18121075 | 2.97 |
ENSDART00000171072
|
SEC14L1
|
si:dkey-237i9.1 |
| chr16_+_19033554 | 2.97 |
ENSDART00000182430
|
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr14_-_8795081 | 2.96 |
ENSDART00000106671
|
tgfb2l
|
transforming growth factor, beta 2, like |
| chr7_-_71829865 | 2.96 |
ENSDART00000168754
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr12_-_3903886 | 2.96 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr24_-_12745222 | 2.91 |
ENSDART00000151836
|
si:ch211-196f5.9
|
si:ch211-196f5.9 |
| chr10_+_26515946 | 2.91 |
ENSDART00000134276
|
synj1
|
synaptojanin 1 |
| chr3_-_50277959 | 2.90 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr14_-_2163454 | 2.89 |
ENSDART00000160123
ENSDART00000169653 |
pcdh2ab9
pcdh2ac
|
protocadherin 2 alpha b 9 protocadherin 2 alpha c |
| chr24_-_27419198 | 2.88 |
ENSDART00000141124
|
ccl34b.4
|
chemokine (C-C motif) ligand 34b, duplicate 4 |
| chr7_-_48665305 | 2.88 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr6_+_54142311 | 2.85 |
ENSDART00000154115
|
hmga1b
|
high mobility group AT-hook 1b |
| chr9_-_28867562 | 2.83 |
ENSDART00000189597
ENSDART00000060321 |
zgc:91818
|
zgc:91818 |
| chr15_+_17446796 | 2.82 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr25_-_2081371 | 2.80 |
ENSDART00000104915
ENSDART00000156925 |
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr21_+_30351256 | 2.80 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr24_-_21923930 | 2.79 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr18_+_49969568 | 2.78 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr16_+_50006145 | 2.77 |
ENSDART00000049375
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr16_+_11483811 | 2.76 |
ENSDART00000169012
ENSDART00000173042 |
grik5
|
glutamate receptor, ionotropic, kainate 5 |
| chr9_+_38883388 | 2.75 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr5_-_28625515 | 2.74 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
Network of associatons between targets according to the STRING database.
First level regulatory network of runx2a+runx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.1 | GO:0015840 | urea transport(GO:0015840) |
| 2.3 | 9.4 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 2.2 | 6.5 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 1.9 | 5.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 1.7 | 12.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 1.5 | 4.6 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.4 | 28.8 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 1.2 | 3.7 | GO:0002858 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.0 | 1.9 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.9 | 11.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.9 | 7.3 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.9 | 3.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.9 | 7.7 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.9 | 2.6 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.8 | 5.6 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.8 | 8.7 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.8 | 3.1 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.8 | 4.7 | GO:0003417 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.8 | 3.8 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.7 | 2.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.7 | 3.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.7 | 4.9 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.7 | 6.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.7 | 7.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.6 | 1.2 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.6 | 1.2 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.6 | 6.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.6 | 3.5 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.6 | 7.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.5 | 5.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.5 | 2.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.5 | 4.5 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.5 | 1.4 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.4 | 2.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.4 | 3.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.4 | 23.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.4 | 1.7 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.4 | 18.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 2.9 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.4 | 2.0 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.4 | 4.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.4 | 1.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.4 | 1.1 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.4 | 1.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 2.6 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.4 | 23.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.4 | 4.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.4 | 1.1 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.3 | 4.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.3 | 1.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.3 | 1.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 0.9 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.3 | 4.6 | GO:0048016 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.3 | 8.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.3 | 2.4 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.3 | 1.7 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 8.3 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.3 | 3.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 0.8 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.3 | 3.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.3 | 1.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 11.2 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.3 | 5.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 2.6 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 1.3 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 0.7 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.2 | 2.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 2.4 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.2 | 0.9 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 0.9 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.2 | 17.2 | GO:0060348 | bone development(GO:0060348) |
| 0.2 | 8.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 0.7 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 1.8 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 4.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 0.6 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 1.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.2 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.0 | GO:0061339 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.2 | 2.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 6.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 1.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 2.0 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 1.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 0.7 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 2.9 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.2 | 4.2 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.2 | 1.8 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.2 | 2.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 1.7 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.2 | 1.9 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.2 | 3.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 3.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.7 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 3.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.8 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) organ maturation(GO:0048799) |
| 0.1 | 5.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.5 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.8 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 2.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.9 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 7.4 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 1.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.7 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.4 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 1.8 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.3 | GO:0021527 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 4.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 3.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 1.1 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 3.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.4 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 9.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.3 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 0.6 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.1 | 24.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.8 | GO:1905066 | canonical Wnt signaling pathway involved in heart development(GO:0061316) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.8 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 1.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 3.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.6 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.1 | 9.4 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.1 | 4.1 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 16.8 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.1 | 0.3 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.3 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.3 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 2.3 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 2.5 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 6.0 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 3.3 | GO:0048596 | embryonic camera-type eye morphogenesis(GO:0048596) |
| 0.1 | 2.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.6 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.1 | 1.1 | GO:0060896 | neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.1 | 3.1 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.6 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 3.5 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.1 | 1.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.7 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 0.7 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 6.3 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.1 | 2.5 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.1 | 0.5 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.8 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.1 | 4.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 2.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 2.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 4.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 3.3 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.9 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.8 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 2.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.9 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 2.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.9 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 2.2 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 2.5 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.0 | 0.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 1.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 3.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.3 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 5.5 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 2.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.5 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.9 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.7 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.5 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.5 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 2.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 3.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.6 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 3.7 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 3.0 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 2.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.5 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.8 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 5.7 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 1.8 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 2.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 2.1 | GO:1902652 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.8 | GO:0040008 | regulation of growth(GO:0040008) |
| 0.0 | 1.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.8 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
| 0.0 | 0.3 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.6 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 1.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 1.9 | GO:0030509 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.5 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 1.4 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.9 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 2.7 | GO:0040007 | growth(GO:0040007) |
| 0.0 | 1.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.0 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 3.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0003205 | cardiac chamber development(GO:0003205) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.3 | GO:0060271 | cilium morphogenesis(GO:0060271) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 63.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 1.1 | 4.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.8 | 3.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.7 | 7.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.6 | 8.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.6 | 4.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.5 | 5.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.5 | 3.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.5 | 3.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.5 | 1.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 5.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 13.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.4 | 1.7 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.4 | 7.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 3.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.4 | 1.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 1.9 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.4 | 5.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 7.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 1.7 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 2.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 6.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 4.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.3 | 9.8 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.3 | 3.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 7.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 3.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.2 | 3.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 0.7 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 2.6 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 2.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.2 | 4.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 3.1 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.2 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 2.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.7 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 1.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 1.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.4 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 8.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 4.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 6.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 4.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 1.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 0.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 17.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 1.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 7.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 6.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 7.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 6.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 5.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 3.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.3 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 5.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.4 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 12.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 2.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.0 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 6.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 8.4 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.9 | GO:0098858 | actin-based cell projection(GO:0098858) |
| 0.0 | 3.7 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 2.9 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.8 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.1 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 28.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.8 | 5.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.5 | 4.5 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 1.5 | 14.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 1.3 | 3.9 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 1.1 | 5.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 1.0 | 4.2 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 1.0 | 6.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.8 | 3.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.8 | 4.6 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.8 | 3.8 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.8 | 4.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 3.5 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.7 | 4.9 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.6 | 3.8 | GO:0097363 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.6 | 3.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.6 | 3.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.6 | 27.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.6 | 1.7 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.5 | 3.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.5 | 2.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.5 | 3.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 1.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.4 | 1.3 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.4 | 5.6 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.4 | 5.5 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.4 | 1.5 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 2.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 1.8 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.4 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 4.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 3.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 2.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 5.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 2.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 8.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 5.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.3 | 2.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 4.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.3 | 6.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 4.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 4.6 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.3 | 5.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.3 | 2.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 1.1 | GO:0033745 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.3 | 2.7 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.3 | 11.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 3.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.3 | 3.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 8.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 0.9 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 3.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 4.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.1 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.6 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.2 | 4.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.2 | 3.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 1.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 2.2 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 2.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 6.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 3.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 9.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 10.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 2.0 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 0.9 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 2.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 0.7 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 2.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.2 | 2.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 3.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 5.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 4.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 6.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 2.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 10.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 4.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.1 | 1.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 8.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 4.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.1 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 3.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 3.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 3.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 6.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 26.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 1.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 25.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.4 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 3.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 2.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 5.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 3.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 12.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 1.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 9.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 2.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 3.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 6.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.4 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 2.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.5 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 3.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.9 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 1.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.7 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 1.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 9.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.9 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.7 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.1 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 3.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 2.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.0 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 3.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 23.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 4.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 8.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 2.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 5.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 5.9 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 8.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 3.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 4.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 6.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 6.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 9.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 11.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.5 | 8.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 6.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 4.5 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.5 | 6.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 4.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.4 | 27.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.3 | 4.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 3.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 3.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 3.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.3 | 1.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 1.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 3.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 2.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.3 | 3.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 3.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 4.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 2.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 1.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 5.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 3.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.1 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.0 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.9 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 3.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |