Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for runx1
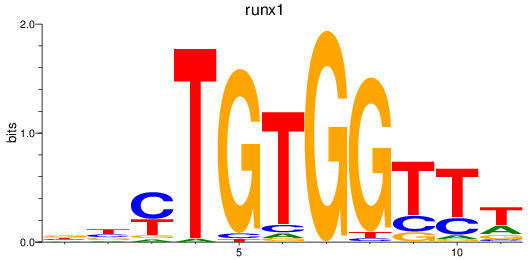
Z-value: 1.99
Transcription factors associated with runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx1
|
ENSDARG00000087646 | RUNX family transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx1 | dr11_v1_chr1_-_1402303_1402303 | 0.27 | 9.2e-03 | Click! |
Activity profile of runx1 motif
Sorted Z-values of runx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_7810348 | 24.60 |
ENSDART00000171984
|
cxcl19
|
chemokine (C-X-C motif) ligand 19 |
| chr17_-_2039511 | 21.89 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr18_+_5543677 | 20.01 |
ENSDART00000146161
ENSDART00000136189 |
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr7_+_22637515 | 19.93 |
ENSDART00000158698
|
si:dkey-112a7.5
|
si:dkey-112a7.5 |
| chr14_+_38786298 | 19.72 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr20_-_14054083 | 19.33 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr7_+_31879986 | 17.28 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr23_+_36653376 | 16.63 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr3_-_16784280 | 16.24 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr7_+_31879649 | 15.97 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr19_-_40191358 | 15.87 |
ENSDART00000183919
|
grn1
|
granulin 1 |
| chr5_+_57658898 | 15.81 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr16_+_11724230 | 15.34 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr8_+_47099033 | 15.05 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr8_+_19356072 | 14.44 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr22_+_34616151 | 14.02 |
ENSDART00000155399
ENSDART00000104705 |
si:ch1073-214b20.2
|
si:ch1073-214b20.2 |
| chr18_+_35742838 | 13.51 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
| chr4_+_77973876 | 12.76 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr17_-_8862424 | 12.66 |
ENSDART00000064633
|
nkl.4
|
NK-lysin tandem duplicate 4 |
| chr15_-_12011390 | 12.50 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr1_-_38195012 | 12.05 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr24_-_25244637 | 11.77 |
ENSDART00000153798
|
hhla2b.2
|
HERV-H LTR-associating 2b, tandem duplicate 2 |
| chr1_+_55140970 | 11.68 |
ENSDART00000039807
|
mb
|
myoglobin |
| chr9_+_23770666 | 11.46 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr10_+_17088261 | 11.44 |
ENSDART00000132103
|
si:dkey-106l3.7
|
si:dkey-106l3.7 |
| chr7_+_24523017 | 11.35 |
ENSDART00000077047
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr4_-_20043484 | 11.20 |
ENSDART00000167780
|
zgc:193726
|
zgc:193726 |
| chr5_-_30615901 | 11.19 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr19_-_7144548 | 11.09 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr4_-_77557279 | 10.97 |
ENSDART00000180113
|
AL935186.10
|
|
| chr19_-_40192249 | 10.67 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chr18_+_31073265 | 10.63 |
ENSDART00000023539
|
cyba
|
cytochrome b-245, alpha polypeptide |
| chr19_+_5640504 | 10.60 |
ENSDART00000179987
|
ft2
|
alpha(1,3)fucosyltransferase gene 2 |
| chr2_-_37465517 | 10.48 |
ENSDART00000139983
|
si:dkey-57k2.6
|
si:dkey-57k2.6 |
| chr17_-_8899323 | 10.46 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr11_+_24703108 | 10.34 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr7_-_17337233 | 10.29 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr13_-_39947335 | 9.97 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr11_+_21076872 | 9.88 |
ENSDART00000155521
|
prelp
|
proline/arginine-rich end leucine-rich repeat protein |
| chr4_-_13502549 | 9.75 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr5_+_13373593 | 9.67 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr1_-_34335752 | 9.66 |
ENSDART00000140157
|
si:dkey-24h22.5
|
si:dkey-24h22.5 |
| chr6_-_39764995 | 9.58 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr10_-_15128771 | 9.52 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr4_-_68563862 | 9.41 |
ENSDART00000182970
|
BX548011.2
|
|
| chr17_+_1627379 | 9.36 |
ENSDART00000184050
|
LO018432.1
|
|
| chr22_+_21255860 | 9.34 |
ENSDART00000134893
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr10_+_43994471 | 9.34 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr22_-_31020690 | 9.33 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr25_-_3470910 | 9.15 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr1_-_12027 | 9.15 |
ENSDART00000164359
|
rpl24
|
ribosomal protein L24 |
| chr1_-_57629639 | 9.11 |
ENSDART00000158984
|
zmp:0000001289
|
zmp:0000001289 |
| chr16_-_17056630 | 9.10 |
ENSDART00000138715
|
plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr15_-_23376541 | 9.06 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr14_-_11430566 | 8.88 |
ENSDART00000137154
ENSDART00000091158 |
irg1l
|
immunoresponsive gene 1, like |
| chr11_-_30630628 | 8.87 |
ENSDART00000103265
|
zgc:158773
|
zgc:158773 |
| chr10_-_43771447 | 8.86 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr8_+_21353878 | 8.85 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr2_+_32796873 | 8.85 |
ENSDART00000077511
|
ccr9a
|
chemokine (C-C motif) receptor 9a |
| chr24_+_19518570 | 8.71 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr19_-_3821678 | 8.70 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr13_-_33170733 | 8.66 |
ENSDART00000057382
|
fbln5
|
fibulin 5 |
| chr19_-_5372572 | 8.62 |
ENSDART00000151326
|
krt17
|
keratin 17 |
| chr9_+_30464641 | 8.60 |
ENSDART00000128357
|
gja5a
|
gap junction protein, alpha 5a |
| chr1_+_55703120 | 8.57 |
ENSDART00000141089
|
adgre6
|
adhesion G protein-coupled receptor E6 |
| chr3_-_34022076 | 8.55 |
ENSDART00000150907
ENSDART00000151092 |
ighv1-4
ighv1-4
|
immunoglobulin heavy variable 1-4 immunoglobulin heavy variable 1-4 |
| chr1_+_54650048 | 8.52 |
ENSDART00000141207
|
si:ch211-202h22.7
|
si:ch211-202h22.7 |
| chr25_+_4751879 | 8.48 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr25_-_13202208 | 8.46 |
ENSDART00000168155
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr2_+_51028269 | 8.44 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr25_-_23526058 | 8.37 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr14_+_30279391 | 8.33 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr13_-_20381485 | 8.32 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr7_+_39706004 | 8.30 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr3_+_3598555 | 8.22 |
ENSDART00000191152
|
CR589947.3
|
|
| chr17_+_6563307 | 8.20 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr6_-_15096556 | 8.17 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr22_-_5323482 | 8.12 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr13_-_25305179 | 8.08 |
ENSDART00000110064
|
plaua
|
plasminogen activator, urokinase a |
| chr18_+_22109379 | 8.05 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr6_-_43449013 | 8.00 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr24_-_25098719 | 7.98 |
ENSDART00000193651
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr4_-_16353733 | 7.92 |
ENSDART00000186785
|
lum
|
lumican |
| chr12_+_34273240 | 7.90 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr7_+_22691961 | 7.90 |
ENSDART00000041615
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr21_+_11248448 | 7.87 |
ENSDART00000142431
|
arid6
|
AT-rich interaction domain 6 |
| chr19_+_3849378 | 7.86 |
ENSDART00000166218
ENSDART00000159228 |
oscp1a
|
organic solute carrier partner 1a |
| chr10_+_24692076 | 7.85 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr13_+_25396896 | 7.84 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr23_-_37575030 | 7.84 |
ENSDART00000031875
|
tor1l3
|
torsin family 1 like 3 |
| chr22_+_22004082 | 7.82 |
ENSDART00000148375
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr17_-_6382392 | 7.81 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr2_-_40199780 | 7.81 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr15_+_36054864 | 7.80 |
ENSDART00000156697
|
col4a3
|
collagen, type IV, alpha 3 |
| chr21_+_22985078 | 7.70 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr4_-_12795436 | 7.63 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr22_-_8860869 | 7.54 |
ENSDART00000166188
|
CABZ01046427.2
|
|
| chr20_+_98179 | 7.43 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr13_-_37122217 | 7.39 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr1_-_9228007 | 7.38 |
ENSDART00000147277
ENSDART00000135219 |
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr11_-_39202915 | 7.34 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr14_+_22022441 | 7.33 |
ENSDART00000149121
|
clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr9_+_6578580 | 7.29 |
ENSDART00000061577
|
fhl2a
|
four and a half LIM domains 2a |
| chr21_-_11632403 | 7.29 |
ENSDART00000171708
ENSDART00000138619 ENSDART00000136308 ENSDART00000144770 |
cast
|
calpastatin |
| chr1_+_12231478 | 7.27 |
ENSDART00000111485
|
tmod1
|
tropomodulin 1 |
| chr22_-_30770751 | 7.24 |
ENSDART00000172115
|
AL831726.2
|
|
| chr19_-_35428815 | 7.20 |
ENSDART00000169006
ENSDART00000003167 |
ak2
|
adenylate kinase 2 |
| chr21_-_44081540 | 7.20 |
ENSDART00000130833
|
FO704810.1
|
|
| chr8_-_49207319 | 7.19 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr23_-_36316352 | 7.18 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr4_-_26108053 | 7.17 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr23_+_10347851 | 7.11 |
ENSDART00000127667
|
krt18
|
keratin 18 |
| chr16_-_22194400 | 7.08 |
ENSDART00000186042
|
il6r
|
interleukin 6 receptor |
| chr3_-_21382491 | 6.98 |
ENSDART00000016562
|
itga2b
|
integrin, alpha 2b |
| chr7_-_52417060 | 6.98 |
ENSDART00000148579
|
myzap
|
myocardial zonula adherens protein |
| chr14_+_34501245 | 6.93 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr5_-_39474235 | 6.92 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr15_+_12435975 | 6.89 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr10_+_2742499 | 6.87 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr3_-_29870848 | 6.82 |
ENSDART00000186457
|
rpl3
|
ribosomal protein L3 |
| chr2_-_16159491 | 6.78 |
ENSDART00000110059
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr1_-_26702930 | 6.75 |
ENSDART00000109297
ENSDART00000152389 |
foxe1
|
forkhead box E1 |
| chr17_-_10838434 | 6.75 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr1_-_9227804 | 6.67 |
ENSDART00000190360
|
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr21_-_226071 | 6.67 |
ENSDART00000160667
|
nup54
|
nucleoporin 54 |
| chr15_-_25571865 | 6.65 |
ENSDART00000077836
|
mmp20b
|
matrix metallopeptidase 20b (enamelysin) |
| chr7_+_49681040 | 6.64 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr16_-_30885838 | 6.61 |
ENSDART00000131356
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr24_+_11334733 | 6.60 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr6_-_29007493 | 6.57 |
ENSDART00000065139
|
gfi1ab
|
growth factor independent 1A transcription repressor b |
| chr4_-_13518381 | 6.55 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr7_-_2039060 | 6.47 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr19_+_37118547 | 6.47 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr10_+_35329751 | 6.44 |
ENSDART00000148043
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr14_-_26498196 | 6.44 |
ENSDART00000054175
ENSDART00000145625 ENSDART00000183347 ENSDART00000191084 ENSDART00000191143 |
smad5
|
SMAD family member 5 |
| chr21_-_38852860 | 6.43 |
ENSDART00000166101
|
tlr22
|
toll-like receptor 22 |
| chr12_-_30540699 | 6.43 |
ENSDART00000167712
ENSDART00000102464 |
zgc:153920
|
zgc:153920 |
| chr6_-_7769178 | 6.42 |
ENSDART00000191701
ENSDART00000149823 |
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr2_-_51700709 | 6.41 |
ENSDART00000188601
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr7_+_48761875 | 6.39 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr2_+_36898982 | 6.39 |
ENSDART00000084859
|
rabgap1l2
|
RAB GTPase activating protein 1-like 2 |
| chr15_+_37412883 | 6.38 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr7_-_38658411 | 6.36 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr10_+_38643304 | 6.35 |
ENSDART00000067447
|
mmp30
|
matrix metallopeptidase 30 |
| chr21_+_45685757 | 6.31 |
ENSDART00000160530
|
sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr20_-_23852174 | 6.28 |
ENSDART00000122414
|
si:dkey-15j16.6
|
si:dkey-15j16.6 |
| chr22_-_15578402 | 6.28 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr25_+_29472361 | 6.27 |
ENSDART00000154857
|
il17rel
|
interleukin 17 receptor E-like |
| chr8_+_6576940 | 6.26 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr3_-_7990516 | 6.26 |
ENSDART00000167877
|
TRIM35 (1 of many)
|
si:ch211-175l6.2 |
| chr19_+_19786117 | 6.21 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr2_-_37862380 | 6.17 |
ENSDART00000186005
|
si:ch211-284o19.8
|
si:ch211-284o19.8 |
| chr22_+_19264181 | 6.16 |
ENSDART00000192750
ENSDART00000137591 ENSDART00000136197 |
si:dkey-78l4.5
si:dkey-21e2.11
|
si:dkey-78l4.5 si:dkey-21e2.11 |
| chr2_-_51719439 | 6.15 |
ENSDART00000170385
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr11_+_27347076 | 6.11 |
ENSDART00000173383
|
fbln2
|
fibulin 2 |
| chr20_+_46216431 | 6.10 |
ENSDART00000185384
|
stx7l
|
syntaxin 7-like |
| chr9_-_23990416 | 6.10 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr1_-_9109699 | 6.10 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr7_-_3894831 | 6.05 |
ENSDART00000172921
|
si:dkey-88n24.11
|
si:dkey-88n24.11 |
| chr3_-_7948799 | 6.05 |
ENSDART00000163714
|
trim35-24
|
tripartite motif containing 35-24 |
| chr2_-_16159203 | 6.02 |
ENSDART00000153480
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr23_+_35847538 | 6.02 |
ENSDART00000143935
|
rarga
|
retinoic acid receptor gamma a |
| chr1_-_7951002 | 6.01 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr7_+_48761646 | 6.01 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr21_-_28439596 | 6.00 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr3_-_48259289 | 5.99 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr18_+_3037998 | 5.98 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr3_-_19367081 | 5.96 |
ENSDART00000191369
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr19_+_4068134 | 5.94 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr9_-_14055959 | 5.90 |
ENSDART00000146675
|
fer1l6
|
fer-1-like family member 6 |
| chr10_+_9550419 | 5.90 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr3_-_32590164 | 5.88 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr21_+_25198637 | 5.86 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr20_-_51399133 | 5.86 |
ENSDART00000151426
|
si:ch73-91k6.2
|
si:ch73-91k6.2 |
| chr3_+_22335030 | 5.85 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr5_-_23909934 | 5.83 |
ENSDART00000142516
|
si:ch211-135f11.1
|
si:ch211-135f11.1 |
| chr17_+_5976683 | 5.83 |
ENSDART00000110276
|
zgc:194275
|
zgc:194275 |
| chr15_-_33965440 | 5.80 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr6_-_2162446 | 5.80 |
ENSDART00000171265
|
tgm5l
|
transglutaminase 5, like |
| chr19_+_4062101 | 5.79 |
ENSDART00000166773
|
btr25
|
bloodthirsty-related gene family, member 25 |
| chr11_+_13524504 | 5.68 |
ENSDART00000139889
|
arrdc2
|
arrestin domain containing 2 |
| chr6_+_45932276 | 5.62 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr23_-_4975452 | 5.61 |
ENSDART00000105241
ENSDART00000169978 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr8_+_22377936 | 5.60 |
ENSDART00000181548
|
si:dkey-23c22.5
|
si:dkey-23c22.5 |
| chr9_-_40765868 | 5.56 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr25_+_3104959 | 5.56 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr14_+_15597049 | 5.56 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr9_-_9419704 | 5.55 |
ENSDART00000138996
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr21_+_11560153 | 5.55 |
ENSDART00000065842
|
cd8a
|
CD8a molecule |
| chr3_-_32958505 | 5.54 |
ENSDART00000147374
ENSDART00000136919 |
casp6l1
|
caspase 6, apoptosis-related cysteine peptidase, like 1 |
| chr11_+_24313931 | 5.54 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr9_-_3496548 | 5.54 |
ENSDART00000102876
|
cybrd1
|
cytochrome b reductase 1 |
| chr22_-_10117918 | 5.53 |
ENSDART00000177475
ENSDART00000128590 |
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr16_-_11859309 | 5.53 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr16_-_51254694 | 5.52 |
ENSDART00000148894
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr9_+_9296222 | 5.51 |
ENSDART00000141325
|
si:ch211-214p13.8
|
si:ch211-214p13.8 |
| chr4_+_73224617 | 5.49 |
ENSDART00000187014
ENSDART00000165194 |
CABZ01021435.1
|
|
| chr1_-_59360114 | 5.44 |
ENSDART00000160445
|
CABZ01052573.1
|
|
| chr16_+_1383914 | 5.42 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr13_-_18174864 | 5.39 |
ENSDART00000079884
|
alox5a
|
arachidonate 5-lipoxygenase a |
Network of associatons between targets according to the STRING database.
First level regulatory network of runx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 33.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 5.0 | 20.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 4.8 | 19.3 | GO:0015840 | urea transport(GO:0015840) |
| 3.3 | 10.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 3.2 | 12.8 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 3.0 | 9.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 2.2 | 6.7 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 2.2 | 8.7 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 2.1 | 4.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 2.1 | 23.3 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 2.1 | 12.4 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 2.0 | 12.0 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 1.8 | 5.4 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 1.7 | 13.9 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 1.7 | 5.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 1.6 | 3.3 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 1.6 | 7.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 1.4 | 11.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 1.4 | 5.5 | GO:0010039 | response to iron ion(GO:0010039) |
| 1.4 | 6.8 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 1.3 | 4.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 1.3 | 6.6 | GO:0097186 | amelogenesis(GO:0097186) |
| 1.3 | 6.4 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 1.3 | 14.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 1.3 | 8.8 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 1.3 | 8.8 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 1.2 | 25.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 1.1 | 4.5 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 1.1 | 3.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 1.1 | 5.5 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.1 | 3.3 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 1.1 | 8.7 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 1.1 | 3.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 1.0 | 7.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 1.0 | 21.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 1.0 | 3.0 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 1.0 | 7.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 1.0 | 2.9 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 1.0 | 2.9 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 1.0 | 15.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.9 | 4.7 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.9 | 5.5 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.9 | 80.0 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.9 | 8.1 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.9 | 9.6 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.8 | 5.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.8 | 4.1 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.8 | 2.4 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.8 | 7.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.8 | 3.2 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.8 | 5.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.8 | 6.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.8 | 11.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.8 | 15.8 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.7 | 3.0 | GO:0009265 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.7 | 7.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.7 | 7.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.7 | 5.6 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.7 | 9.6 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.7 | 4.7 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.7 | 2.7 | GO:0042755 | eating behavior(GO:0042755) |
| 0.6 | 1.3 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.6 | 3.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.6 | 1.9 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.6 | 2.5 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.6 | 13.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.6 | 4.9 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.6 | 1.2 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.6 | 1.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.6 | 1.8 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.6 | 2.4 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) positive regulation of lipid catabolic process(GO:0050996) |
| 0.6 | 2.3 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.6 | 6.9 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.6 | 2.9 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.6 | 3.4 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.6 | 3.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.6 | 4.0 | GO:1900117 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.6 | 5.6 | GO:2000403 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.6 | 1.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.5 | 3.8 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.5 | 7.6 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.5 | 3.8 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.5 | 71.8 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.5 | 2.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.5 | 4.0 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.5 | 6.0 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.5 | 2.5 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.5 | 2.9 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.5 | 2.9 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.5 | 1.4 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.5 | 7.2 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.5 | 1.9 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.5 | 5.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.5 | 2.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.5 | 4.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.5 | 3.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.5 | 2.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.5 | 0.9 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.4 | 4.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.4 | 2.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.4 | 1.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.4 | 8.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.4 | 6.6 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.4 | 3.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.4 | 1.6 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 8.8 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.4 | 9.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.4 | 12.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.4 | 3.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.4 | 1.2 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.4 | 3.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.4 | 35.5 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.4 | 4.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 | 0.4 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.4 | 1.1 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.4 | 35.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.4 | 4.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.4 | 1.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 1.4 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 10.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 3.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 5.1 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.3 | 4.7 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.3 | 2.0 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 15.5 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.3 | 2.2 | GO:0000730 | DNA recombinase assembly(GO:0000730) |
| 0.3 | 1.0 | GO:0034398 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.3 | 1.6 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.3 | 6.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 2.8 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.3 | 4.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.3 | 9.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 6.0 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.3 | 2.0 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.3 | 1.4 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.3 | 3.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 0.8 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 3.0 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 1.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 1.6 | GO:0070935 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.3 | 3.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.3 | 2.6 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.3 | 12.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.3 | 3.5 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 6.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.2 | 5.2 | GO:0007210 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) serotonin receptor signaling pathway(GO:0007210) |
| 0.2 | 10.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.2 | 6.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 0.9 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.2 | 6.6 | GO:0002685 | regulation of leukocyte migration(GO:0002685) |
| 0.2 | 3.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 3.9 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.2 | 26.0 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.2 | 7.0 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.2 | 1.9 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 3.1 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 3.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 0.6 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 9.4 | GO:0060348 | bone development(GO:0060348) |
| 0.2 | 2.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.2 | 9.1 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.2 | 3.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.2 | 2.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 1.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 2.3 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.2 | 2.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 4.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 2.0 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.2 | 4.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 3.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 4.2 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 0.7 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 2.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 10.2 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.2 | 3.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.2 | 3.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.2 | 1.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.2 | 0.3 | GO:0001658 | ureteric bud development(GO:0001657) branching involved in ureteric bud morphogenesis(GO:0001658) mesonephros development(GO:0001823) ureteric bud morphogenesis(GO:0060675) regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) mesonephric tubule morphogenesis(GO:0072171) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.2 | 4.0 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 2.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 2.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 1.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 6.5 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.2 | 2.8 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.2 | 2.9 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 1.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.2 | 4.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.2 | 0.8 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.6 | GO:0060547 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 7.5 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.1 | 2.8 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 1.4 | GO:0065005 | plasma lipoprotein particle assembly(GO:0034377) protein-lipid complex assembly(GO:0065005) |
| 0.1 | 0.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 4.3 | GO:0019915 | lipid storage(GO:0019915) |
| 0.1 | 1.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 2.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.9 | GO:0006501 | macromitophagy(GO:0000423) C-terminal protein lipidation(GO:0006501) |
| 0.1 | 7.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 3.2 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.1 | 7.7 | GO:0002768 | immune response-regulating cell surface receptor signaling pathway(GO:0002768) |
| 0.1 | 1.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 3.0 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 1.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.7 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 2.5 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 1.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.9 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 2.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 7.3 | GO:0007492 | endoderm development(GO:0007492) |
| 0.1 | 0.9 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.5 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.1 | 0.4 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 6.7 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.1 | 2.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.2 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.1 | 5.2 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.1 | 1.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 1.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.5 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.6 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 1.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 1.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.2 | GO:0001573 | ganglioside metabolic process(GO:0001573) ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 6.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 6.6 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 14.2 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.7 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 0.5 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 0.4 | GO:0070586 | cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 2.5 | GO:0045089 | positive regulation of defense response(GO:0031349) positive regulation of innate immune response(GO:0045089) |
| 0.1 | 0.7 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 2.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 2.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 13.5 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 1.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 6.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 3.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.3 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.1 | 1.1 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 1.0 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.6 | GO:0042493 | response to drug(GO:0042493) |
| 0.1 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 1.9 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 1.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 2.1 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.7 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 2.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.7 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.1 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0009204 | nucleoside triphosphate catabolic process(GO:0009143) deoxyribonucleoside triphosphate catabolic process(GO:0009204) |
| 0.0 | 2.9 | GO:0032273 | positive regulation of protein polymerization(GO:0032273) |
| 0.0 | 8.7 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 1.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 2.1 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.5 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 1.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:1904590 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.0 | 1.4 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.4 | GO:0051984 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.4 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.1 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 1.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 2.0 | GO:0050657 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 1.2 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 2.0 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.6 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.2 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 1.9 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 1.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 2.0 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 2.1 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.9 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.4 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.1 | GO:0031646 | positive regulation of neurological system process(GO:0031646) |
| 0.0 | 0.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.8 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.1 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 1.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) erythrocyte homeostasis(GO:0034101) |
| 0.0 | 1.1 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 1.5 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 2.8 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 1.1 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 13.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.4 | 5.6 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 1.4 | 5.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.3 | 3.9 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 1.3 | 7.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.1 | 7.4 | GO:0016234 | inclusion body(GO:0016234) |
| 1.0 | 2.9 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.9 | 8.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.9 | 13.7 | GO:0070187 | telosome(GO:0070187) |
| 0.9 | 6.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.9 | 9.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.8 | 6.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.8 | 3.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.8 | 2.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.8 | 7.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.7 | 2.2 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.7 | 4.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.7 | 8.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.6 | 5.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.5 | 2.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.5 | 2.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.5 | 3.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.5 | 1.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.5 | 2.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 1.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 1.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.4 | 2.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.4 | 1.7 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.4 | 1.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 11.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.4 | 2.8 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.4 | 24.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 1.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.4 | 2.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.4 | 1.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 1.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 17.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.3 | 6.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 2.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 65.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.3 | 1.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.3 | 15.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 0.8 | GO:0030689 | Noc complex(GO:0030689) |
| 0.3 | 4.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 3.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 9.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 1.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 10.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.2 | 11.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 1.4 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.2 | 11.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 19.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 3.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 8.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 2.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 2.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 3.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 4.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 2.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 162.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 13.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.4 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 1.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 6.8 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 1.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 1.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 7.3 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 21.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 4.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 12.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 9.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 9.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.0 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 5.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 15.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.0 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 7.5 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 1.8 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 7.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 5.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 1.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 6.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 9.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 1.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 1.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 13.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 2.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.0 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0000793 | condensed chromosome(GO:0000793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 20.0 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 3.0 | 33.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 2.9 | 8.8 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 2.6 | 7.8 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 2.5 | 9.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 2.3 | 18.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 2.2 | 24.6 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.1 | 10.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 2.1 | 18.5 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 1.8 | 5.5 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 1.7 | 1.7 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 1.5 | 4.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 1.5 | 4.4 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.4 | 6.8 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 1.3 | 7.8 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 1.3 | 25.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 1.3 | 3.9 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 1.3 | 15.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 1.2 | 2.4 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.1 | 3.3 | GO:0042806 | fucose binding(GO:0042806) |
| 1.1 | 7.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 1.0 | 3.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.0 | 7.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.0 | 4.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 1.0 | 9.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.0 | 11.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.0 | 2.9 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) |
| 1.0 | 3.9 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.9 | 5.4 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.9 | 3.6 | GO:0008329 | lipopolysaccharide receptor activity(GO:0001875) signaling pattern recognition receptor activity(GO:0008329) |
| 0.9 | 2.7 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.9 | 9.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.9 | 2.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 17.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.8 | 2.5 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.8 | 2.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.8 | 3.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.8 | 19.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.8 | 9.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.7 | 2.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.7 | 3.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.7 | 2.1 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.7 | 2.8 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.7 | 3.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.7 | 8.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.6 | 4.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.6 | 11.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 13.3 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.6 | 8.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.6 | 3.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.6 | 4.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.6 | 7.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.6 | 4.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.6 | 11.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.6 | 2.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.6 | 1.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.6 | 3.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 63.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 4.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 7.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.5 | 3.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.5 | 4.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 3.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.5 | 3.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 2.9 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.5 | 21.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.5 | 1.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.5 | 2.8 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 1.4 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.5 | 15.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.5 | 1.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.5 | 1.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 1.3 | GO:0032404 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.4 | 3.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 9.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.4 | 2.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 27.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.4 | 5.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 2.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 12.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 4.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.4 | 3.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.4 | 1.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.3 | 1.4 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.3 | 20.5 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.3 | 2.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.3 | 4.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.3 | 17.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 7.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 1.3 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.3 | 4.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 1.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 6.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 2.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.3 | 1.6 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.3 | 3.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.3 | 2.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 0.9 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.3 | 2.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.3 | 2.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 2.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 2.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 10.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 1.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.3 | 18.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 3.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 2.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.3 | 5.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.3 | 15.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 18.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.3 | 5.0 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 1.5 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.2 | 8.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 8.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.2 | 6.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.7 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.2 | 10.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 2.0 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.6 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 77.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.2 | 3.5 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.2 | 0.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 5.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 2.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 15.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 1.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 2.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.9 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 0.9 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.2 | 3.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.2 | 35.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 2.3 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 2.8 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 1.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 4.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 2.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 36.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 2.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 3.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 3.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 23.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 0.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 18.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 2.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 0.8 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 3.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 2.8 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 5.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 13.3 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 4.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 10.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 3.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.4 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.1 | 2.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 6.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 12.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 2.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 7.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 1.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 3.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.9 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 5.5 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 0.7 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 2.3 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 6.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.1 | 1.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 2.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 4.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.1 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 3.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 2.0 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 5.8 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.1 | 4.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.5 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.5 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 20.9 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 5.2 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 11.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 8.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.7 | 13.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.7 | 6.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.7 | 17.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.6 | 16.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.6 | 17.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.5 | 4.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.5 | 6.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.5 | 14.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.5 | 2.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.4 | 2.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 3.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.4 | 6.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 3.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 1.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 2.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.3 | 5.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.3 | 6.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 13.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 13.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.3 | 8.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.3 | 4.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 2.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.3 | 39.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 10.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.3 | 5.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.3 | 5.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 3.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 2.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 2.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 2.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 3.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 3.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 30.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.2 | 2.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 4.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 7.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.2 | 26.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 1.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.2 | 2.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 3.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 2.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 23.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 1.5 | 7.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.9 | 4.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.8 | 6.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.8 | 15.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.7 | 7.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.7 | 9.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.7 | 5.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.7 | 6.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.6 | 2.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.6 | 6.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.6 | 8.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.6 | 10.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.5 | 3.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.5 | 6.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.5 | 9.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.5 | 6.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.5 | 5.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.5 | 1.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.5 | 7.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.5 | 5.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 3.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 3.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 2.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.4 | 8.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 15.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 35.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 3.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 0.9 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.3 | 2.9 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.3 | 2.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 13.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 3.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 1.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.3 | 3.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 1.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.3 | 7.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 2.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 4.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 8.5 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 3.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 2.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 4.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 3.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 10.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 4.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.2 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 4.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.9 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 2.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 3.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |