Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
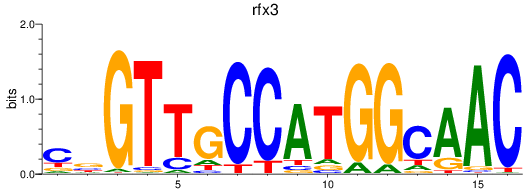
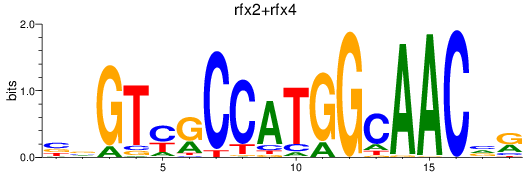
Results for rfx3_rfx2+rfx4
Z-value: 4.31
Transcription factors associated with rfx3_rfx2+rfx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx3
|
ENSDARG00000014550 | regulatory factor X, 3 (influences HLA class II expression) |
|
rfx2
|
ENSDARG00000013575 | regulatory factor X, 2 (influences HLA class II expression) |
|
rfx4
|
ENSDARG00000026395 | regulatory factor X, 4 |
|
rfx4
|
ENSDARG00000116861 | regulatory factor X, 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx4 | dr11_v1_chr18_-_15373620_15373620 | 0.79 | 4.8e-21 | Click! |
| rfx3 | dr11_v1_chr10_-_641609_641609 | 0.49 | 4.5e-07 | Click! |
| rfx2 | dr11_v1_chr8_-_20138054_20138251 | -0.13 | 2.0e-01 | Click! |
Activity profile of rfx3_rfx2+rfx4 motif
Sorted Z-values of rfx3_rfx2+rfx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_26697536 | 61.23 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr19_-_9472893 | 55.81 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr20_-_9436521 | 52.27 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr17_-_6759006 | 46.03 |
ENSDART00000184692
ENSDART00000180530 |
vsnl1b
|
visinin-like 1b |
| chr24_-_24163201 | 42.43 |
ENSDART00000140170
|
map7d2b
|
MAP7 domain containing 2b |
| chr23_-_19953089 | 40.88 |
ENSDART00000153828
|
atp2b3b
|
ATPase plasma membrane Ca2+ transporting 3b |
| chr5_+_32162684 | 39.43 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr5_-_23317477 | 38.88 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
| chr10_-_35542071 | 38.85 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr13_-_36911118 | 38.73 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr16_-_13662514 | 38.73 |
ENSDART00000146348
|
shisa7a
|
shisa family member 7a |
| chr21_-_43952958 | 36.64 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr16_+_221739 | 36.50 |
ENSDART00000180243
|
nrsn1
|
neurensin 1 |
| chr11_-_97817 | 36.20 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr6_-_33023745 | 35.72 |
ENSDART00000156211
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr7_-_58098814 | 35.24 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr20_-_34868814 | 34.89 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr5_-_21044693 | 33.99 |
ENSDART00000140298
|
si:dkey-13n15.2
|
si:dkey-13n15.2 |
| chr3_-_1190132 | 33.85 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr17_-_14726824 | 33.33 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr7_-_52842605 | 33.25 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr21_-_15929041 | 32.98 |
ENSDART00000080693
|
lhx5
|
LIM homeobox 5 |
| chr5_-_29643930 | 32.95 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr24_-_24162930 | 32.82 |
ENSDART00000080602
|
map7d2b
|
MAP7 domain containing 2b |
| chr9_-_31747106 | 32.40 |
ENSDART00000048469
ENSDART00000145204 ENSDART00000186889 |
nalcn
|
sodium leak channel, non-selective |
| chr23_+_35708730 | 32.13 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr1_+_54908895 | 32.12 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr20_+_20637866 | 32.11 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr13_+_28821841 | 31.65 |
ENSDART00000179900
|
CU639469.1
|
|
| chr18_+_7283283 | 31.50 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr25_-_13381854 | 31.46 |
ENSDART00000164621
ENSDART00000169129 |
ndrg4
|
NDRG family member 4 |
| chr13_-_31470439 | 29.92 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr20_+_20638034 | 29.83 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr22_+_16418688 | 29.72 |
ENSDART00000009360
|
ankrd29
|
ankyrin repeat domain 29 |
| chr6_+_13933464 | 28.87 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr13_+_33117528 | 28.79 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr25_-_25736958 | 28.03 |
ENSDART00000166308
|
cib2
|
calcium and integrin binding family member 2 |
| chr7_-_52876683 | 27.83 |
ENSDART00000172910
|
map1aa
|
microtubule-associated protein 1Aa |
| chr24_-_205275 | 27.56 |
ENSDART00000108762
|
vopp1
|
VOPP1, WBP1/VOPP1 family member |
| chr17_+_8988374 | 27.44 |
ENSDART00000109573
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr17_+_15534815 | 26.73 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr19_-_103289 | 26.69 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr19_-_8604429 | 26.31 |
ENSDART00000151165
|
trim46b
|
tripartite motif containing 46b |
| chr18_+_50890749 | 26.01 |
ENSDART00000174109
|
si:ch1073-450f2.1
|
si:ch1073-450f2.1 |
| chr9_-_44289636 | 25.89 |
ENSDART00000110411
|
cerkl
|
ceramide kinase-like |
| chr3_-_6767440 | 25.88 |
ENSDART00000156174
|
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr21_+_41743493 | 25.03 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr5_+_20148671 | 24.31 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr9_-_3671911 | 24.10 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr3_-_52644737 | 23.56 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr3_-_50277959 | 23.52 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr20_+_591505 | 23.44 |
ENSDART00000046438
|
kcnk2b
|
potassium channel, subfamily K, member 2b |
| chr11_-_29650930 | 23.31 |
ENSDART00000166969
|
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr3_+_31945604 | 23.25 |
ENSDART00000114957
|
zgc:193811
|
zgc:193811 |
| chr4_-_8903240 | 23.21 |
ENSDART00000129983
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr14_-_25577094 | 23.03 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr17_+_33296436 | 22.41 |
ENSDART00000123114
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr2_+_18988407 | 22.16 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr22_+_31930650 | 21.73 |
ENSDART00000092090
|
dock3
|
dedicator of cytokinesis 3 |
| chr2_+_47581997 | 21.40 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr18_+_50275933 | 21.26 |
ENSDART00000143911
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr24_-_6678640 | 21.12 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr18_-_46763170 | 21.06 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr16_-_12984631 | 21.00 |
ENSDART00000184863
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr19_+_43123331 | 20.65 |
ENSDART00000187836
|
CABZ01101996.1
|
|
| chr22_-_25033105 | 20.36 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr5_+_64732036 | 20.36 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr10_+_37145007 | 20.28 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr21_-_22709251 | 20.13 |
ENSDART00000140032
|
si:dkeyp-69c1.9
|
si:dkeyp-69c1.9 |
| chr5_+_20147830 | 20.07 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr2_-_1569250 | 19.98 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr17_-_40397752 | 19.91 |
ENSDART00000178483
|
BX548062.1
|
|
| chr1_+_45085194 | 19.82 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr12_-_41684729 | 19.79 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr1_-_56223913 | 19.77 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr16_+_27957808 | 19.67 |
ENSDART00000133696
|
znf804b
|
zinc finger protein 804B |
| chr15_-_47892583 | 19.61 |
ENSDART00000192584
|
DMWD
|
zmp:0000000529 |
| chr16_-_9869056 | 19.60 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr8_-_7504146 | 18.87 |
ENSDART00000148339
ENSDART00000140098 |
cdk20
|
cyclin-dependent kinase 20 |
| chr18_-_21271373 | 18.86 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr10_-_17232372 | 18.65 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr23_+_19558574 | 18.61 |
ENSDART00000137811
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr5_+_66353750 | 18.49 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr9_+_2393764 | 18.48 |
ENSDART00000172624
|
chn1
|
chimerin 1 |
| chr16_-_44399335 | 18.43 |
ENSDART00000165058
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr3_-_34337969 | 18.31 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr5_-_57920600 | 18.25 |
ENSDART00000132376
|
si:dkey-27p23.3
|
si:dkey-27p23.3 |
| chr2_+_47582681 | 18.16 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr16_+_4497302 | 17.76 |
ENSDART00000081826
ENSDART00000148096 |
ttc29
|
tetratricopeptide repeat domain 29 |
| chr5_+_66353589 | 17.55 |
ENSDART00000138246
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr5_-_46273938 | 17.51 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr23_-_28692581 | 17.34 |
ENSDART00000078141
|
ribc1
|
RIB43A domain with coiled-coils 1 |
| chr10_-_17231917 | 17.29 |
ENSDART00000038577
|
rab36
|
RAB36, member RAS oncogene family |
| chr15_-_19669770 | 17.27 |
ENSDART00000152707
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr17_-_43558494 | 17.09 |
ENSDART00000103830
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr17_+_33296852 | 17.05 |
ENSDART00000154580
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr18_+_50276337 | 16.93 |
ENSDART00000140352
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr10_+_2234283 | 16.79 |
ENSDART00000136363
|
cntnap3
|
contactin associated protein like 3 |
| chr2_-_21167652 | 16.74 |
ENSDART00000185792
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr6_-_21582444 | 16.41 |
ENSDART00000151339
|
si:dkey-43k4.3
|
si:dkey-43k4.3 |
| chr16_-_50157187 | 16.11 |
ENSDART00000155162
|
nek10
|
NIMA-related kinase 10 |
| chr2_-_44038698 | 15.87 |
ENSDART00000079582
ENSDART00000146804 |
kirrel1b
|
kirre like nephrin family adhesion molecule 1b |
| chr18_+_11970987 | 15.87 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr11_+_45299447 | 15.86 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr5_-_30704390 | 15.85 |
ENSDART00000016709
|
ift22
|
intraflagellar transport 22 homolog (Chlamydomonas) |
| chr2_-_36819624 | 15.78 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr14_-_52542928 | 15.58 |
ENSDART00000075749
|
ppp2r2ba
|
protein phosphatase 2, regulatory subunit B, beta a |
| chr24_+_744713 | 15.49 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr14_-_9355177 | 15.36 |
ENSDART00000138535
|
fam46d
|
family with sequence similarity 46, member D |
| chr3_-_12026741 | 15.36 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr20_-_28404362 | 14.98 |
ENSDART00000055932
ENSDART00000188161 |
pigh
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr25_+_25737386 | 14.97 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr17_-_45009782 | 14.90 |
ENSDART00000123971
|
fam161b
|
family with sequence similarity 161, member B |
| chr4_+_4902392 | 14.82 |
ENSDART00000133866
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr3_+_33367954 | 14.76 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr13_-_6218248 | 14.63 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr1_-_45633955 | 14.62 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr3_-_12890670 | 14.59 |
ENSDART00000159934
ENSDART00000188607 |
btbd17b
|
BTB (POZ) domain containing 17b |
| chr7_-_72423666 | 14.53 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr17_+_28882977 | 14.38 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr11_+_29770966 | 14.36 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr9_+_54891962 | 14.25 |
ENSDART00000148620
ENSDART00000105620 |
si:ch211-12n8.3
|
si:ch211-12n8.3 |
| chr9_+_23748342 | 14.12 |
ENSDART00000019053
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr20_+_20672163 | 14.07 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr8_-_19216657 | 14.07 |
ENSDART00000135096
ENSDART00000135869 ENSDART00000145951 |
si:ch73-222f22.2
si:ch73-222f22.2
|
si:ch73-222f22.2 si:ch73-222f22.2 |
| chr3_-_19899914 | 14.07 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr16_+_24632096 | 14.01 |
ENSDART00000157237
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr5_-_69271450 | 13.96 |
ENSDART00000133802
|
inpp5jb
|
inositol polyphosphate-5-phosphatase Jb |
| chr10_+_25222367 | 13.87 |
ENSDART00000042767
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr5_-_64213253 | 13.81 |
ENSDART00000171711
|
gpsm1a
|
G protein signaling modulator 1a |
| chr3_+_60716904 | 13.77 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr17_+_14425219 | 13.71 |
ENSDART00000153994
|
ccdc175
|
coiled-coil domain containing 175 |
| chr8_-_49345388 | 13.64 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr17_+_33767890 | 13.63 |
ENSDART00000193177
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr6_+_58543336 | 13.63 |
ENSDART00000157018
|
stmn3
|
stathmin-like 3 |
| chr10_+_36037977 | 13.63 |
ENSDART00000164678
|
katnal1
|
katanin p60 subunit A-like 1 |
| chr17_+_3390772 | 13.52 |
ENSDART00000160073
|
sntg2
|
syntrophin, gamma 2 |
| chr4_-_27398385 | 13.48 |
ENSDART00000142117
ENSDART00000150553 ENSDART00000182746 |
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr2_+_35728033 | 13.46 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr17_-_39772999 | 13.34 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr21_-_35325466 | 13.25 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr6_+_10534752 | 13.18 |
ENSDART00000090874
|
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr24_-_32408404 | 13.03 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr24_+_26345427 | 12.92 |
ENSDART00000089756
|
lrrc34
|
leucine rich repeat containing 34 |
| chr17_-_24684687 | 12.80 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr3_-_22829710 | 12.73 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr23_-_20133994 | 12.73 |
ENSDART00000004871
|
lrrc23
|
leucine rich repeat containing 23 |
| chr16_-_45069882 | 12.71 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr6_-_40744720 | 12.59 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr25_+_35375848 | 12.51 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr24_+_17262879 | 12.51 |
ENSDART00000145949
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr8_-_19216180 | 12.44 |
ENSDART00000146162
|
si:ch73-222f22.2
|
si:ch73-222f22.2 |
| chr8_-_39859688 | 12.36 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr18_-_41232297 | 12.23 |
ENSDART00000036928
|
fbxo36a
|
F-box protein 36a |
| chr4_+_31259 | 12.23 |
ENSDART00000166826
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr10_-_36682509 | 12.19 |
ENSDART00000148093
ENSDART00000063365 |
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr8_+_23809843 | 12.16 |
ENSDART00000099724
|
si:ch211-163l21.10
|
si:ch211-163l21.10 |
| chr12_-_47793857 | 11.97 |
ENSDART00000161294
|
dydc2
|
DPY30 domain containing 2 |
| chr8_-_18229169 | 11.90 |
ENSDART00000131764
ENSDART00000143036 ENSDART00000145986 |
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr5_-_42661012 | 11.90 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr3_+_50312422 | 11.88 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr1_-_45647846 | 11.84 |
ENSDART00000186881
|
BX511120.1
|
|
| chr10_+_34685135 | 11.83 |
ENSDART00000184999
|
nbeaa
|
neurobeachin a |
| chr19_+_22062202 | 11.82 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr5_+_42386705 | 11.74 |
ENSDART00000143034
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_+_42407962 | 11.74 |
ENSDART00000188489
|
BX548073.11
|
|
| chr17_-_39779906 | 11.67 |
ENSDART00000155181
|
pimr61
|
Pim proto-oncogene, serine/threonine kinase, related 61 |
| chr9_+_23748133 | 11.63 |
ENSDART00000180758
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr24_-_1021318 | 11.48 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr5_-_23179319 | 11.43 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr13_+_33304187 | 11.41 |
ENSDART00000075826
ENSDART00000145295 |
dcdc2b
|
doublecortin domain containing 2B |
| chr5_-_38506981 | 11.40 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr23_+_22335407 | 11.37 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr2_-_14271874 | 11.36 |
ENSDART00000189390
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr3_-_30123113 | 11.34 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr19_-_9712530 | 11.34 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr13_-_36703164 | 11.33 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr14_-_46395659 | 11.26 |
ENSDART00000083833
|
bbs7
|
Bardet-Biedl syndrome 7 |
| chr9_-_25255490 | 11.25 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr8_+_47571211 | 11.24 |
ENSDART00000131460
|
plch2a
|
phospholipase C, eta 2a |
| chr18_+_30028637 | 11.20 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr25_+_37480285 | 11.19 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr24_+_26345609 | 11.09 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
| chr21_-_22557469 | 11.09 |
ENSDART00000167230
|
cfap53
|
cilia and flagella associated protein 53 |
| chr3_-_37351225 | 11.05 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr25_+_17862338 | 11.05 |
ENSDART00000151853
ENSDART00000090867 |
btbd10a
|
BTB (POZ) domain containing 10a |
| chr9_-_16853462 | 10.90 |
ENSDART00000160273
|
CT573248.2
|
|
| chr21_-_43550120 | 10.85 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr12_+_25600685 | 10.66 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr3_-_23596809 | 10.57 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr8_-_25814263 | 10.53 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr7_+_7279903 | 10.41 |
ENSDART00000102603
|
si:ch211-168d1.3
|
si:ch211-168d1.3 |
| chr6_-_13308813 | 10.29 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr2_+_47582488 | 10.24 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr4_-_12388535 | 10.16 |
ENSDART00000017180
|
rergla
|
RERG/RAS-like a |
| chr15_-_1534232 | 10.11 |
ENSDART00000056763
ENSDART00000133943 |
ift80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr4_-_9586713 | 10.10 |
ENSDART00000145613
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr11_+_36477481 | 10.09 |
ENSDART00000128245
|
ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr5_-_71705191 | 10.02 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr17_+_28883353 | 9.99 |
ENSDART00000110322
|
prkd1
|
protein kinase D1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx3_rfx2+rfx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.9 | GO:2000425 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 7.0 | 35.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 7.0 | 21.1 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 6.7 | 26.7 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 6.7 | 26.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 6.3 | 18.8 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 5.2 | 52.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 4.6 | 18.5 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 4.3 | 38.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 4.1 | 24.4 | GO:1901490 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 3.9 | 15.7 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 3.9 | 27.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 3.7 | 55.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 3.5 | 38.9 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 3.2 | 16.0 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 3.2 | 22.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 2.7 | 13.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 2.5 | 25.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 2.5 | 20.0 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 2.5 | 14.8 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 2.3 | 61.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 2.3 | 25.8 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 2.3 | 48.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 2.3 | 34.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 2.3 | 15.9 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 2.3 | 20.4 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 2.2 | 31.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.2 | 26.7 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 2.2 | 15.1 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 2.1 | 21.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 2.1 | 29.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 2.0 | 33.9 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.9 | 5.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.8 | 33.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 1.8 | 12.7 | GO:0032475 | otolith formation(GO:0032475) |
| 1.8 | 5.4 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 1.8 | 7.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 1.7 | 14.0 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 1.7 | 5.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.6 | 4.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 1.5 | 7.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 1.5 | 9.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 1.4 | 10.0 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 1.4 | 12.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 1.4 | 21.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 1.3 | 4.0 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 1.3 | 12.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 1.3 | 17.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 1.3 | 14.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 1.3 | 12.5 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 1.2 | 49.8 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 1.2 | 36.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 1.2 | 15.8 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 1.2 | 2.4 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 1.1 | 4.6 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.1 | 18.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 1.1 | 8.9 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 1.1 | 15.6 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 1.0 | 18.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.9 | 5.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.9 | 13.6 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.9 | 32.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.9 | 13.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.8 | 4.2 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.8 | 5.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.8 | 3.1 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.8 | 10.8 | GO:0030317 | sperm motility(GO:0030317) |
| 0.8 | 19.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.8 | 35.7 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.8 | 11.4 | GO:0001964 | startle response(GO:0001964) |
| 0.7 | 11.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.7 | 7.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.7 | 3.4 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.7 | 6.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.7 | 12.4 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.6 | 19.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.6 | 4.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.6 | 5.7 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.6 | 4.6 | GO:0036372 | opsin transport(GO:0036372) |
| 0.6 | 10.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.5 | 23.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.5 | 8.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.5 | 9.0 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.5 | 29.9 | GO:0003146 | heart jogging(GO:0003146) |
| 0.5 | 8.8 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.5 | 11.4 | GO:0032474 | otolith morphogenesis(GO:0032474) neuromast hair cell development(GO:0035675) |
| 0.5 | 24.3 | GO:0003341 | cilium movement(GO:0003341) |
| 0.5 | 6.1 | GO:0035094 | response to nicotine(GO:0035094) response to alkaloid(GO:0043279) |
| 0.5 | 10.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.5 | 7.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.5 | 2.9 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.5 | 4.3 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.5 | 2.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.4 | 9.5 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.4 | 18.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.4 | 11.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.4 | 13.8 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.4 | 24.1 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.4 | 5.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.4 | 5.8 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.4 | 23.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.4 | 4.8 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.4 | 12.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.4 | 8.8 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.3 | 19.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.3 | 3.3 | GO:1902686 | positive regulation of mitochondrial membrane permeability(GO:0035794) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.3 | 9.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.3 | 16.6 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.3 | 2.0 | GO:0060827 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.3 | 1.6 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.3 | 14.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.3 | 1.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 2.1 | GO:0072178 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.3 | 8.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 1.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 58.1 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.3 | 10.4 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.2 | 15.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.2 | 1.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 3.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 16.1 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.2 | 7.1 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.2 | 44.4 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.2 | 2.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 8.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.2 | 9.8 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.2 | 14.1 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.2 | 2.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 0.9 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 0.9 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.2 | 71.3 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.2 | 38.3 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.2 | 1.4 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 3.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 4.0 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.2 | 3.0 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 11.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 8.0 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 | 2.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 11.5 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 70.0 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.1 | 8.3 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.1 | 19.4 | GO:0060026 | convergent extension(GO:0060026) |
| 0.1 | 9.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 9.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 8.6 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.1 | 14.1 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 8.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 0.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 11.5 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 3.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 6.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 9.8 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.1 | 7.0 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 2.1 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 11.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 2.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.9 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 1.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 7.0 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.1 | 4.8 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.1 | 24.6 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.1 | 2.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 3.1 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 41.3 | GO:0007010 | cytoskeleton organization(GO:0007010) |
| 0.1 | 5.7 | GO:0050808 | synapse organization(GO:0050808) |
| 0.1 | 5.3 | GO:0030900 | forebrain development(GO:0030900) |
| 0.0 | 0.5 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 3.0 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 8.0 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 4.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 5.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 5.5 | GO:0009116 | nucleoside metabolic process(GO:0009116) |
| 0.0 | 1.7 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0006813 | potassium ion transport(GO:0006813) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 61.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 3.2 | 19.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 3.1 | 18.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 3.0 | 24.1 | GO:0034464 | BBSome(GO:0034464) |
| 2.9 | 8.8 | GO:0001534 | radial spoke(GO:0001534) |
| 2.6 | 33.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 2.4 | 38.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 2.3 | 9.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 2.2 | 8.8 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 2.2 | 35.2 | GO:0030315 | T-tubule(GO:0030315) |
| 2.2 | 33.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 2.1 | 32.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 2.1 | 21.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 2.0 | 35.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 1.8 | 5.4 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 1.6 | 34.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 1.6 | 16.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 1.5 | 9.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 1.3 | 5.2 | GO:0042641 | actomyosin(GO:0042641) |
| 1.3 | 40.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.2 | 29.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 1.1 | 20.2 | GO:0097546 | ciliary base(GO:0097546) |
| 1.1 | 8.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 1.1 | 33.1 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 1.0 | 39.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 1.0 | 23.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 1.0 | 127.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.9 | 68.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.8 | 3.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.7 | 12.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.6 | 1.9 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.6 | 13.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.6 | 4.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.6 | 8.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.6 | 6.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.5 | 25.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.5 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.5 | 8.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 2.9 | GO:0060091 | kinocilium(GO:0060091) |
| 0.5 | 18.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.4 | 27.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.4 | 15.1 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.4 | 3.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 104.5 | GO:0005929 | cilium(GO:0005929) |
| 0.3 | 11.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 3.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 12.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.3 | 13.1 | GO:0030496 | midbody(GO:0030496) |
| 0.3 | 86.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.2 | 19.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 13.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 8.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 14.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 14.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.2 | 16.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 81.3 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.2 | 2.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 51.6 | GO:0045202 | synapse(GO:0045202) |
| 0.2 | 14.2 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.2 | 11.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 18.3 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.2 | 97.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.2 | 1.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 14.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 3.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 71.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 0.8 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 81.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 10.3 | GO:0005938 | cell cortex(GO:0005938) cytoplasmic region(GO:0099568) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 15.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 12.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 22.7 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.3 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 3.7 | 25.9 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 3.2 | 12.7 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 3.2 | 22.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 2.9 | 61.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 2.8 | 19.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 2.7 | 18.8 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 2.2 | 15.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 2.1 | 18.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 2.0 | 78.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.9 | 35.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 1.7 | 14.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 1.7 | 15.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.6 | 33.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.6 | 23.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 1.5 | 38.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 1.4 | 7.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 1.4 | 13.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.4 | 40.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 1.4 | 13.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 1.3 | 4.0 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 1.2 | 31.1 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 1.2 | 13.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.2 | 29.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 1.2 | 6.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.1 | 4.6 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 1.1 | 10.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.1 | 12.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 1.1 | 11.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 1.1 | 4.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.0 | 58.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 1.0 | 5.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.0 | 12.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.9 | 5.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.9 | 24.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.9 | 42.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.8 | 49.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.8 | 5.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.8 | 16.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.8 | 17.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.8 | 3.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.8 | 36.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.7 | 10.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.7 | 34.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.7 | 7.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.7 | 2.1 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.7 | 25.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.7 | 3.4 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.6 | 6.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.6 | 9.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.6 | 7.9 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.5 | 68.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.5 | 8.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.5 | 15.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.5 | 8.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 6.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 155.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.4 | 38.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.4 | 3.3 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.4 | 128.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.4 | 5.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 8.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.3 | 12.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 5.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 18.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.3 | 11.2 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 2.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 9.2 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.3 | 1.9 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 1.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 2.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 6.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 10.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 8.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 3.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 9.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 13.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.3 | 23.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 21.9 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.2 | 2.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.2 | 11.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.2 | 8.3 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.2 | 9.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.2 | 9.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 68.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.2 | 73.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.2 | 5.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 50.6 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.2 | 3.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 19.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.2 | 7.1 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 8.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.1 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 11.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 6.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 3.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 20.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 3.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 8.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 9.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.9 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 7.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 20.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 13.5 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.1 | 9.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 9.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 6.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 40.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 92.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 32.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 4.5 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 3.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 26.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.0 | 25.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.9 | 36.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.8 | 27.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.4 | 6.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.4 | 10.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 11.7 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 11.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 6.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 4.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.2 | 6.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 3.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 9.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 4.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.2 | 36.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 3.6 | 61.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 3.4 | 58.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 2.0 | 32.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.4 | 12.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 1.1 | 19.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.8 | 4.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.8 | 24.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.8 | 8.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.7 | 6.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.7 | 15.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.7 | 8.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.7 | 16.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.6 | 21.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.6 | 5.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.5 | 10.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 8.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 10.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 17.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 35.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 2.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 4.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 8.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |