Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for rest
Z-value: 6.65
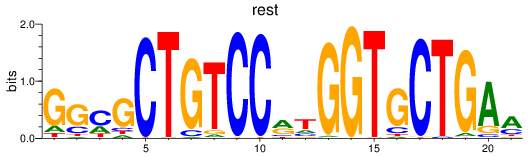
Transcription factors associated with rest
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rest
|
ENSDARG00000103046 | Danio rerio RE1-silencing transcription factor (rest), mRNA. |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rest | dr11_v1_chr14_-_52433292_52433292 | -0.23 | 2.6e-02 | Click! |
Activity profile of rest motif
Sorted Z-values of rest motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_34913069 | 225.34 |
ENSDART00000007584
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr17_-_12385308 | 180.15 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr25_-_22639133 | 171.20 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr9_-_34915351 | 170.95 |
ENSDART00000100728
ENSDART00000139608 |
upf3a
|
UPF3A, regulator of nonsense mediated mRNA decay |
| chr21_+_11468934 | 152.82 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr25_-_19443421 | 150.07 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr20_+_27020201 | 146.99 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr19_-_19339285 | 146.92 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr4_-_5302866 | 146.71 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr5_-_29643930 | 144.40 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr7_+_40228422 | 133.27 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr5_-_29643381 | 131.57 |
ENSDART00000034849
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr23_+_45579497 | 128.95 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr7_+_26224211 | 128.74 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr20_+_45741566 | 128.56 |
ENSDART00000113454
|
chgb
|
chromogranin B |
| chr18_+_26337869 | 126.74 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr20_+_38724575 | 124.97 |
ENSDART00000015095
ENSDART00000152972 |
uts1
|
urotensin 1 |
| chr6_+_41255485 | 124.94 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr25_+_35250976 | 123.48 |
ENSDART00000003494
|
slc17a6a
|
solute carrier family 17 (vesicular glutamate transporter), member 6a |
| chr7_+_23907692 | 121.92 |
ENSDART00000045479
|
syt4
|
synaptotagmin IV |
| chr3_+_33340939 | 121.88 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr6_-_30210378 | 121.69 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr15_+_36156986 | 117.59 |
ENSDART00000059791
|
sst1.1
|
somatostatin 1, tandem duplicate 1 |
| chr10_+_25219728 | 116.60 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr16_+_46148990 | 115.02 |
ENSDART00000083919
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr21_-_42100471 | 114.05 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr10_+_21588002 | 113.26 |
ENSDART00000100596
|
pcdh1a6
|
protocadherin 1 alpha 6 |
| chr21_+_11468642 | 109.18 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr4_-_5302162 | 109.11 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr5_-_29750377 | 108.45 |
ENSDART00000051474
|
barhl1a
|
BarH-like homeobox 1a |
| chr25_-_31863374 | 108.41 |
ENSDART00000028338
|
scamp5a
|
secretory carrier membrane protein 5a |
| chr10_+_23022263 | 108.22 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr13_+_23282549 | 105.14 |
ENSDART00000101134
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr7_-_52876683 | 102.96 |
ENSDART00000172910
|
map1aa
|
microtubule-associated protein 1Aa |
| chr23_+_28693278 | 102.71 |
ENSDART00000078148
|
smc1a
|
structural maintenance of chromosomes 1A |
| chr1_+_54911458 | 102.34 |
ENSDART00000089603
|
golga7ba
|
golgin A7 family, member Ba |
| chr3_+_33341640 | 101.19 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr5_-_26093945 | 101.15 |
ENSDART00000010199
ENSDART00000145096 |
fam219ab
|
family with sequence similarity 219, member Ab |
| chr19_-_43552252 | 98.82 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
| chr20_+_19212962 | 97.69 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr1_+_10720294 | 97.21 |
ENSDART00000139387
|
atp1b1b
|
ATPase Na+/K+ transporting subunit beta 1b |
| chr19_-_42244358 | 95.98 |
ENSDART00000102697
|
cart4
|
cocaine- and amphetamine-regulated transcript 4 |
| chr18_+_28988373 | 93.17 |
ENSDART00000018685
|
syt9a
|
synaptotagmin IXa |
| chr12_+_24561832 | 93.03 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr10_+_21807497 | 91.08 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr14_-_2264494 | 90.93 |
ENSDART00000191149
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr12_+_24342303 | 90.62 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr17_+_19630272 | 89.40 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr3_-_37476475 | 88.82 |
ENSDART00000148107
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr7_-_32833153 | 86.73 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr5_-_32092856 | 86.29 |
ENSDART00000086181
ENSDART00000181677 |
cabp7b
|
calcium binding protein 7b |
| chr7_-_69521481 | 86.02 |
ENSDART00000148465
|
slc1a1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr23_-_17657348 | 85.09 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr23_+_35759843 | 80.62 |
ENSDART00000047082
|
gdap1l1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr7_-_30087048 | 80.12 |
ENSDART00000112743
|
nmbb
|
neuromedin Bb |
| chr23_+_41800052 | 79.04 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr21_+_6751760 | 78.46 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr22_-_25033105 | 76.45 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr18_-_44611252 | 75.88 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr7_+_17229980 | 73.61 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr6_+_43903209 | 72.56 |
ENSDART00000006435
|
gpr27
|
G protein-coupled receptor 27 |
| chr21_-_17482465 | 72.29 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr19_+_26718074 | 71.59 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr10_-_36927594 | 69.07 |
ENSDART00000162046
|
sez6a
|
seizure related 6 homolog a |
| chr9_-_27649406 | 68.40 |
ENSDART00000181270
ENSDART00000187112 |
stxbp5l
|
syntaxin binding protein 5-like |
| chr6_-_43092175 | 67.89 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr14_-_2267515 | 66.54 |
ENSDART00000180988
ENSDART00000130712 |
pcdh2aa15
|
protocadherin 2 alpha a 15 |
| chr21_+_6751405 | 66.40 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr10_-_26793448 | 65.92 |
ENSDART00000141955
|
mcf2b
|
MCF.2 cell line derived transforming sequence b |
| chr1_-_38538284 | 61.81 |
ENSDART00000009777
|
glra3
|
glycine receptor, alpha 3 |
| chr14_-_2032753 | 61.66 |
ENSDART00000111335
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr7_+_59739164 | 59.17 |
ENSDART00000110412
|
gpr78b
|
G protein-coupled receptor 78b |
| chr20_-_35052464 | 58.71 |
ENSDART00000037195
|
kif26bb
|
kinesin family member 26Bb |
| chr13_+_25846528 | 56.46 |
ENSDART00000087426
|
bcl11aa
|
B cell CLL/lymphoma 11Aa |
| chr9_-_31915423 | 54.71 |
ENSDART00000060051
|
fgf14
|
fibroblast growth factor 14 |
| chr17_+_19630068 | 53.64 |
ENSDART00000182619
|
rgs7a
|
regulator of G protein signaling 7a |
| chr14_-_2193248 | 52.46 |
ENSDART00000128659
|
pcdh2ab10
|
protocadherin 2 alpha b 10 |
| chr11_+_19780717 | 48.95 |
ENSDART00000157394
|
cadpsa
|
Ca2+-dependent activator protein for secretion a |
| chr7_-_31940590 | 48.08 |
ENSDART00000131009
|
bdnf
|
brain-derived neurotrophic factor |
| chr15_+_24212847 | 47.11 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr12_+_19305390 | 46.92 |
ENSDART00000183987
ENSDART00000066391 |
csnk1e
|
casein kinase 1, epsilon |
| chr17_-_10249095 | 45.75 |
ENSDART00000159963
|
sstr1a
|
somatostatin receptor 1a |
| chr23_-_30785382 | 43.99 |
ENSDART00000136156
ENSDART00000131285 |
myt1a
|
myelin transcription factor 1a |
| chr17_-_33552363 | 42.21 |
ENSDART00000154400
|
tmem121aa
|
transmembrane protein 121Aa |
| chr14_-_2004291 | 40.92 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr12_-_19862912 | 40.79 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr10_-_29892486 | 40.18 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr2_-_5776030 | 37.73 |
ENSDART00000133851
|
nyap2b
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2b |
| chr6_-_31336317 | 36.07 |
ENSDART00000193927
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr14_-_1987121 | 35.76 |
ENSDART00000135733
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr7_+_17229282 | 35.38 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr11_+_19781053 | 34.96 |
ENSDART00000029680
ENSDART00000182743 |
cadpsa
|
Ca2+-dependent activator protein for secretion a |
| chr20_+_41640687 | 32.30 |
ENSDART00000138686
|
fam184a
|
family with sequence similarity 184, member A |
| chr11_+_33426510 | 30.83 |
ENSDART00000163106
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr14_-_1998501 | 28.22 |
ENSDART00000189052
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr18_+_50089000 | 25.97 |
ENSDART00000058936
|
scamp5b
|
secretory carrier membrane protein 5b |
| chr10_+_36441124 | 25.02 |
ENSDART00000185626
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr23_+_30859086 | 23.45 |
ENSDART00000053661
|
zgc:198371
|
zgc:198371 |
| chr14_-_32486757 | 23.25 |
ENSDART00000148830
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr14_-_1963369 | 23.11 |
ENSDART00000125521
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr15_-_22147860 | 21.91 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr6_+_21740672 | 21.55 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr18_+_36806545 | 21.18 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr17_-_21617618 | 18.31 |
ENSDART00000109031
|
gpr26
|
G protein-coupled receptor 26 |
| chr15_-_47841259 | 17.15 |
ENSDART00000155384
|
kbtbd3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr11_+_33426254 | 15.81 |
ENSDART00000111193
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr1_-_201004 | 14.63 |
ENSDART00000188956
|
FO681286.1
|
|
| chr1_+_12348213 | 12.78 |
ENSDART00000144920
ENSDART00000138759 ENSDART00000067082 |
clta
|
clathrin, light chain A |
| chr13_-_45811137 | 9.40 |
ENSDART00000190512
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr16_-_40727455 | 8.75 |
ENSDART00000162331
|
si:dkey-22o22.2
|
si:dkey-22o22.2 |
| chr4_-_17391091 | 8.37 |
ENSDART00000056002
|
th2
|
tyrosine hydroxylase 2 |
| chr4_+_357810 | 6.62 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr20_-_39391833 | 5.94 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr3_-_44753325 | 5.65 |
ENSDART00000074543
|
hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr3_-_20721064 | 5.04 |
ENSDART00000193532
|
spop
|
speckle-type POZ protein |
| chr12_-_16524727 | 2.73 |
ENSDART00000166645
|
FO704724.1
|
|
| chr1_+_40794395 | 2.52 |
ENSDART00000111656
|
gpr78a
|
G protein-coupled receptor 78a |
| chr4_-_76488581 | 2.16 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr1_+_19515228 | 0.10 |
ENSDART00000103091
|
clrn2
|
clarin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rest
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 49.0 | 147.0 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 38.9 | 116.6 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 29.9 | 538.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 26.3 | 210.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 22.8 | 114.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 21.0 | 146.9 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 18.8 | 150.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 16.2 | 405.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 16.1 | 208.9 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 14.1 | 56.5 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 13.6 | 109.0 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 12.6 | 75.9 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 10.1 | 223.1 | GO:0007631 | feeding behavior(GO:0007631) |
| 8.7 | 121.7 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 8.5 | 67.9 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 7.2 | 36.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 5.9 | 88.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 5.7 | 40.2 | GO:0021982 | pineal gland development(GO:0021982) |
| 5.7 | 170.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 5.5 | 21.9 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 5.4 | 21.6 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 5.1 | 97.2 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 5.0 | 80.1 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 5.0 | 25.0 | GO:1990173 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 4.8 | 61.8 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 4.7 | 215.1 | GO:0050433 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) catecholamine secretion(GO:0050432) regulation of catecholamine secretion(GO:0050433) |
| 4.6 | 45.7 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 4.5 | 40.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 4.2 | 108.4 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 4.0 | 48.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 4.0 | 103.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 4.0 | 102.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 3.9 | 54.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 2.8 | 8.4 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 2.8 | 102.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 2.4 | 612.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 2.4 | 68.4 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 2.1 | 128.6 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 2.0 | 72.6 | GO:0042593 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 1.2 | 105.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 1.2 | 115.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 1.1 | 183.7 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 1.0 | 79.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 1.0 | 178.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.9 | 46.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.4 | 22.5 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.4 | 133.3 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.4 | 117.6 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.4 | 12.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 58.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.2 | 96.3 | GO:0009968 | negative regulation of signal transduction(GO:0009968) |
| 0.2 | 44.0 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.1 | 376.2 | GO:0006357 | regulation of transcription from RNA polymerase II promoter(GO:0006357) |
| 0.1 | 94.5 | GO:0007399 | nervous system development(GO:0007399) |
| 0.0 | 89.2 | GO:0035556 | intracellular signal transduction(GO:0035556) |
| 0.0 | 5.0 | GO:0030162 | regulation of proteolysis(GO:0030162) |
| 0.0 | 26.0 | GO:0015031 | protein transport(GO:0015031) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 67.6 | 405.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 35.9 | 538.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 14.7 | 147.0 | GO:0042583 | chromaffin granule(GO:0042583) |
| 14.0 | 210.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 13.3 | 93.2 | GO:0031045 | dense core granule(GO:0031045) |
| 9.7 | 97.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 8.6 | 102.7 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 7.2 | 21.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 6.8 | 102.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 6.7 | 134.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 5.0 | 208.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 3.7 | 154.8 | GO:0032589 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 3.5 | 252.9 | GO:0030141 | secretory granule(GO:0030141) |
| 2.8 | 116.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 2.4 | 79.0 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 2.3 | 127.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 1.4 | 25.0 | GO:0015030 | Cajal body(GO:0015030) |
| 1.4 | 68.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 1.3 | 121.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 1.2 | 61.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 1.2 | 170.0 | GO:0099503 | secretory vesicle(GO:0099503) |
| 1.1 | 124.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 1.0 | 111.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 1.0 | 21.9 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.7 | 58.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.6 | 232.4 | GO:0045202 | synapse(GO:0045202) |
| 0.5 | 166.1 | GO:0005730 | nucleolus(GO:0005730) |
| 0.5 | 923.5 | GO:0005576 | extracellular region(GO:0005576) |
| 0.4 | 534.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.2 | 5.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 931.3 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.1 | 476.2 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 75.9 | GO:0016020 | membrane(GO:0016020) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 44.6 | 223.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 26.9 | 538.0 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 16.7 | 116.6 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 16.2 | 210.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 13.4 | 80.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 11.4 | 387.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 10.1 | 405.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 8.1 | 114.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 7.8 | 109.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 6.9 | 61.8 | GO:0016934 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 6.1 | 79.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 5.7 | 45.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 5.2 | 170.9 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 5.0 | 105.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 5.0 | 25.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 4.9 | 97.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 4.5 | 215.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 4.0 | 68.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 3.7 | 48.1 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 2.8 | 76.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 2.2 | 242.6 | GO:0005179 | hormone activity(GO:0005179) |
| 2.2 | 102.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 1.7 | 8.4 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 1.0 | 21.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.9 | 12.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.7 | 699.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.7 | 133.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.6 | 5.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.5 | 36.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.5 | 58.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.5 | 86.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.4 | 103.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.4 | 75.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.3 | 197.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.3 | 89.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.3 | 96.6 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.2 | 533.5 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.1 | 121.7 | GO:0005102 | receptor binding(GO:0005102) |
| 0.1 | 394.7 | GO:0046872 | metal ion binding(GO:0046872) |
| 0.1 | 78.2 | GO:0030234 | enzyme regulator activity(GO:0030234) |
| 0.1 | 5.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 140.9 | GO:0005515 | protein binding(GO:0005515) |
| 0.0 | 28.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 126.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 5.0 | 128.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 2.9 | 102.7 | PID ATM PATHWAY | ATM pathway |
| 2.9 | 46.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 1.2 | 48.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.6 | 12.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 21.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 5.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 12.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 31.7 | 126.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 12.3 | 86.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 7.6 | 114.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 7.3 | 109.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 4.4 | 44.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 3.3 | 79.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 3.1 | 102.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 2.1 | 12.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 1.4 | 21.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.3 | 36.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 1.3 | 170.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 1.1 | 46.9 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.4 | 5.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |