Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for rel
Z-value: 1.21
Transcription factors associated with rel
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rel
|
ENSDARG00000055276 | v-rel avian reticuloendotheliosis viral oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rel | dr11_v1_chr13_-_25819825_25819825 | 0.51 | 1.2e-07 | Click! |
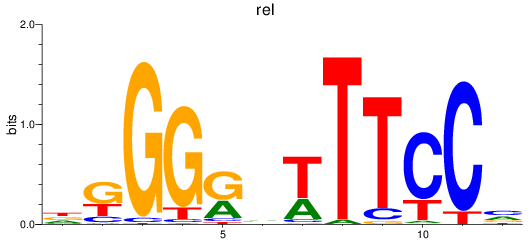
Activity profile of rel motif
Sorted Z-values of rel motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_16780643 | 14.33 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr24_+_26006730 | 14.12 |
ENSDART00000140384
ENSDART00000139184 |
ccl20b
|
chemokine (C-C motif) ligand 20b |
| chr7_+_45975537 | 13.44 |
ENSDART00000170253
|
plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr7_+_12950507 | 13.22 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr1_-_49225890 | 13.03 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr25_-_13188214 | 12.78 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr2_-_51772438 | 11.96 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr22_+_5478353 | 11.59 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr20_+_16881883 | 10.82 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr23_+_19701587 | 10.72 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr1_+_18716 | 10.60 |
ENSDART00000172454
ENSDART00000161190 |
nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr4_-_12795436 | 10.29 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr14_+_36885524 | 10.20 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr3_+_54744069 | 9.47 |
ENSDART00000134958
ENSDART00000114443 |
si:ch211-74m13.3
|
si:ch211-74m13.3 |
| chr16_+_23403602 | 9.10 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr18_-_5527050 | 9.08 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr8_-_18667693 | 8.90 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr22_+_661711 | 8.68 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr2_+_45191049 | 8.21 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr1_-_52447364 | 8.09 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr18_+_30847237 | 7.89 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr17_-_6382392 | 7.73 |
ENSDART00000188051
ENSDART00000192560 ENSDART00000137389 ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr8_+_24281512 | 7.65 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr16_+_38394371 | 7.45 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr6_-_54815886 | 7.45 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr14_+_36889893 | 7.40 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr6_-_436658 | 7.37 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr11_-_28050559 | 7.37 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr13_-_39947335 | 7.37 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr6_+_3334710 | 7.16 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr14_+_20893065 | 7.15 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr4_-_18954001 | 7.13 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr5_-_42878178 | 7.12 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr4_-_12790886 | 7.06 |
ENSDART00000182535
ENSDART00000067131 ENSDART00000186426 |
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr3_+_34670076 | 7.04 |
ENSDART00000133457
|
dlx4a
|
distal-less homeobox 4a |
| chr5_-_30984271 | 6.94 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr19_+_7043634 | 6.91 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr3_-_32590164 | 6.79 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr13_-_25819825 | 6.67 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr14_+_3038473 | 6.50 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr7_-_20241346 | 6.47 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr6_-_52675630 | 6.33 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr14_-_6987649 | 6.33 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr6_-_442163 | 6.27 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr14_-_40821411 | 6.19 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr21_+_43328685 | 6.11 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr13_-_36034582 | 6.05 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr16_-_29714540 | 5.82 |
ENSDART00000067854
|
tnfaip8l2b
|
tumor necrosis factor, alpha-induced protein 8-like 2b |
| chr12_-_30760971 | 5.75 |
ENSDART00000066257
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr22_+_6293563 | 5.71 |
ENSDART00000063416
|
rnasel2
|
ribonuclease like 2 |
| chr5_+_27525477 | 5.62 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr5_+_13373593 | 5.59 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr6_+_3334392 | 5.40 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr20_-_33976937 | 5.32 |
ENSDART00000136834
|
sele
|
selectin E |
| chr8_+_39674707 | 5.28 |
ENSDART00000126301
ENSDART00000040330 |
prkab1b
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit, b |
| chr7_+_39706004 | 5.27 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr1_-_37377509 | 5.18 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr13_+_669177 | 5.16 |
ENSDART00000190085
|
CU462915.1
|
|
| chr7_-_8309505 | 5.15 |
ENSDART00000182530
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr6_+_37655078 | 5.14 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr18_-_49286381 | 5.14 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr12_-_11349899 | 5.11 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
| chr19_+_23298037 | 5.08 |
ENSDART00000183948
|
irgf1
|
immunity-related GTPase family, f1 |
| chr18_-_18543358 | 5.01 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr14_-_40850481 | 4.99 |
ENSDART00000173236
|
elf1
|
E74-like ETS transcription factor 1 |
| chr22_+_8536838 | 4.92 |
ENSDART00000132998
|
si:ch73-27e22.7
|
si:ch73-27e22.7 |
| chr8_-_12432604 | 4.85 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr3_-_21242460 | 4.82 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr16_-_24832038 | 4.45 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr14_-_1646360 | 4.22 |
ENSDART00000186528
|
LO018106.2
|
|
| chr11_+_14333441 | 4.12 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr17_+_12942634 | 4.09 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr16_-_41762983 | 4.07 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr14_+_30773764 | 4.02 |
ENSDART00000186961
|
atl3
|
atlastin 3 |
| chr6_+_28428329 | 4.01 |
ENSDART00000188056
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_8129536 | 3.90 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr25_-_35960229 | 3.85 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr12_-_26430507 | 3.84 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr8_+_22355909 | 3.82 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr7_+_21275152 | 3.72 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr2_+_36939749 | 3.71 |
ENSDART00000005382
|
gadd45bb
|
growth arrest and DNA-damage-inducible, beta b |
| chr18_-_11729 | 3.64 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr22_-_23067859 | 3.53 |
ENSDART00000137873
|
ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr8_+_52642869 | 3.51 |
ENSDART00000163617
ENSDART00000189997 |
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr20_+_23501535 | 3.46 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr2_-_45191319 | 3.46 |
ENSDART00000192272
|
CR407590.2
|
|
| chr1_+_12135129 | 3.45 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr8_+_22955478 | 3.43 |
ENSDART00000099911
|
zgc:136605
|
zgc:136605 |
| chr8_-_26937225 | 3.38 |
ENSDART00000191787
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr3_+_19216567 | 3.37 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr24_+_80653 | 3.33 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr6_+_19933763 | 3.32 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr8_-_13010011 | 3.23 |
ENSDART00000140969
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr1_+_17892944 | 3.21 |
ENSDART00000013021
|
tlr3
|
toll-like receptor 3 |
| chr15_+_27364394 | 3.15 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr3_-_58733718 | 3.10 |
ENSDART00000154603
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr18_+_7073130 | 3.08 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr9_-_5263947 | 3.01 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr16_-_25739331 | 2.97 |
ENSDART00000189455
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr23_-_20361971 | 2.90 |
ENSDART00000133373
|
si:rp71-17i16.5
|
si:rp71-17i16.5 |
| chr20_-_39367895 | 2.84 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr14_-_11529311 | 2.83 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr22_-_651719 | 2.82 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr7_+_29293452 | 2.82 |
ENSDART00000127358
|
si:ch211-112g6.4
|
si:ch211-112g6.4 |
| chr13_+_22717366 | 2.82 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr23_-_33738570 | 2.80 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr13_-_22843562 | 2.79 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr1_+_27690 | 2.73 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr18_-_16924221 | 2.73 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr2_+_2737422 | 2.73 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr16_+_52999778 | 2.73 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr7_+_4384863 | 2.71 |
ENSDART00000042955
ENSDART00000134653 |
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr14_-_33218418 | 2.66 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr14_+_26224731 | 2.64 |
ENSDART00000168673
|
gm2a
|
GM2 ganglioside activator |
| chr1_+_18550864 | 2.64 |
ENSDART00000142515
|
si:dkey-192k22.2
|
si:dkey-192k22.2 |
| chr9_-_18716 | 2.62 |
ENSDART00000164763
|
CABZ01078737.1
|
|
| chr23_-_38054 | 2.60 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr22_+_508290 | 2.60 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr25_+_8955530 | 2.59 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr8_-_46572298 | 2.56 |
ENSDART00000030470
|
sult1st3
|
sulfotransferase family 1, cytosolic sulfotransferase 3 |
| chr20_-_33966148 | 2.55 |
ENSDART00000148111
|
selp
|
selectin P |
| chr14_+_16765992 | 2.51 |
ENSDART00000140061
|
sqstm1
|
sequestosome 1 |
| chr4_+_77681389 | 2.47 |
ENSDART00000099727
|
gbp4
|
guanylate binding protein 4 |
| chr14_+_14806851 | 2.47 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr12_-_19250854 | 2.46 |
ENSDART00000152844
|
cdc42ep1a
|
CDC42 effector protein (Rho GTPase binding) 1a |
| chr7_+_34786591 | 2.46 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr23_+_19606291 | 2.46 |
ENSDART00000139415
|
flnb
|
filamin B, beta (actin binding protein 278) |
| chr20_-_20410029 | 2.45 |
ENSDART00000192177
ENSDART00000063483 |
prkchb
|
protein kinase C, eta, b |
| chr13_+_22717939 | 2.42 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr15_+_6459847 | 2.42 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr15_-_2803313 | 2.41 |
ENSDART00000060839
|
tgfb1a
|
transforming growth factor, beta 1a |
| chr19_-_7043355 | 2.36 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr22_-_31517300 | 2.31 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr4_-_71708855 | 2.26 |
ENSDART00000158819
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr5_+_45139196 | 2.26 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_-_22311610 | 2.23 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr12_+_27462225 | 2.22 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr20_-_51307815 | 2.21 |
ENSDART00000098833
|
nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr16_+_27444098 | 2.18 |
ENSDART00000157690
|
invs
|
inversin |
| chr12_-_3903886 | 2.17 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr19_-_10432134 | 2.10 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr15_+_42235449 | 2.07 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr2_-_53592532 | 1.98 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr11_+_6456146 | 1.97 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr14_-_30967284 | 1.96 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr7_-_3792972 | 1.95 |
ENSDART00000146304
|
si:dkey-28d5.12
|
si:dkey-28d5.12 |
| chr12_-_4206869 | 1.91 |
ENSDART00000106572
|
si:dkey-32n7.9
|
si:dkey-32n7.9 |
| chr15_+_37436430 | 1.91 |
ENSDART00000124779
|
igflr1
|
IGF-like family receptor 1 |
| chr8_+_18101608 | 1.91 |
ENSDART00000140941
|
glis1b
|
GLIS family zinc finger 1b |
| chr21_-_22357545 | 1.90 |
ENSDART00000134320
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr8_-_25771474 | 1.89 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr17_-_10315644 | 1.89 |
ENSDART00000168572
|
ttc6
|
tetratricopeptide repeat domain 6 |
| chr18_-_16922905 | 1.87 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr14_+_6182346 | 1.83 |
ENSDART00000075525
|
si:ch73-22a13.3
|
si:ch73-22a13.3 |
| chr20_+_33981946 | 1.82 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr25_-_7723685 | 1.79 |
ENSDART00000146882
|
phf21ab
|
PHD finger protein 21Ab |
| chr15_+_791220 | 1.79 |
ENSDART00000136137
|
si:dkey-7i4.1
|
si:dkey-7i4.1 |
| chr21_+_21817975 | 1.78 |
ENSDART00000151292
ENSDART00000166704 |
neu3.5
|
sialidase 3 (membrane sialidase), tandem duplicate 5 |
| chr19_+_48351624 | 1.77 |
ENSDART00000169729
|
sgo1
|
shugoshin 1 |
| chr2_+_42191592 | 1.74 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr3_+_16668609 | 1.73 |
ENSDART00000185646
|
zgc:55558
|
zgc:55558 |
| chr5_+_54422941 | 1.72 |
ENSDART00000175157
|
traf2b
|
Tnf receptor-associated factor 2b |
| chr14_+_26224541 | 1.71 |
ENSDART00000128971
|
gm2a
|
GM2 ganglioside activator |
| chr5_+_25596234 | 1.66 |
ENSDART00000143949
ENSDART00000167193 |
si:dkey-96l17.6
|
si:dkey-96l17.6 |
| chr20_-_44496245 | 1.65 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr4_+_75575252 | 1.65 |
ENSDART00000166536
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr22_-_8499778 | 1.64 |
ENSDART00000142219
|
si:ch73-27e22.4
|
si:ch73-27e22.4 |
| chr10_+_40756352 | 1.63 |
ENSDART00000156210
ENSDART00000144576 |
taar19f
|
trace amine associated receptor 19f |
| chr4_+_18806251 | 1.60 |
ENSDART00000138662
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr10_+_22536693 | 1.58 |
ENSDART00000185037
ENSDART00000146040 |
efnb3a
|
ephrin-B3a |
| chr24_+_22731228 | 1.57 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr22_+_18319666 | 1.54 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr17_-_39761086 | 1.51 |
ENSDART00000032410
|
gpr132a
|
G protein-coupled receptor 132a |
| chr13_-_25196758 | 1.50 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr18_-_17077419 | 1.50 |
ENSDART00000148714
|
il17c
|
interleukin 17c |
| chr5_+_37729207 | 1.47 |
ENSDART00000184378
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr22_-_16180467 | 1.45 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr8_-_10949847 | 1.44 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr19_+_28187480 | 1.44 |
ENSDART00000183825
|
irx4b
|
iroquois homeobox 4b |
| chr12_+_4149112 | 1.44 |
ENSDART00000166191
|
itgam
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr7_+_4538334 | 1.43 |
ENSDART00000139326
ENSDART00000146915 |
si:dkey-83f18.9
|
si:dkey-83f18.9 |
| chr2_-_29485408 | 1.42 |
ENSDART00000013411
|
cahz
|
carbonic anhydrase |
| chr11_-_40101246 | 1.40 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr25_-_28926330 | 1.37 |
ENSDART00000155173
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr17_+_8175998 | 1.36 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr22_-_8536482 | 1.36 |
ENSDART00000148134
|
si:ch73-27e22.2
|
si:ch73-27e22.2 |
| chr6_+_46668717 | 1.36 |
ENSDART00000064853
|
oxtrl
|
oxytocin receptor like |
| chr16_-_55259199 | 1.34 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr3_-_55531450 | 1.33 |
ENSDART00000155376
|
tex2
|
testis expressed 2 |
| chr3_-_49815223 | 1.33 |
ENSDART00000181208
|
gcgra
|
glucagon receptor a |
| chr4_-_76488581 | 1.30 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr5_-_22602979 | 1.29 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr11_+_35050253 | 1.27 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr4_+_55059525 | 1.26 |
ENSDART00000128531
|
si:dkey-19c16.12
|
si:dkey-19c16.12 |
| chr9_-_21459074 | 1.26 |
ENSDART00000136427
|
zmym2
|
zinc finger, MYM-type 2 |
| chr7_-_41915312 | 1.20 |
ENSDART00000159869
|
dnaja2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr3_-_31619463 | 1.13 |
ENSDART00000124559
|
moto
|
minamoto |
| chr3_-_54544612 | 1.12 |
ENSDART00000018044
|
angptl6
|
angiopoietin-like 6 |
| chr16_+_54588930 | 1.12 |
ENSDART00000159174
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr4_-_149334 | 1.11 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rel
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.0 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 3.6 | 10.8 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 2.8 | 8.4 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 2.6 | 15.7 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 2.6 | 7.7 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 2.5 | 7.4 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 2.4 | 7.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 2.4 | 7.1 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 1.7 | 5.0 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 1.6 | 14.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 1.6 | 6.3 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 1.6 | 6.3 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 1.3 | 5.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 1.2 | 4.8 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 1.1 | 3.3 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 1.1 | 3.2 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 1.0 | 4.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 1.0 | 5.7 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.9 | 2.8 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.8 | 2.5 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.8 | 45.8 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.8 | 10.3 | GO:0034121 | regulation of toll-like receptor signaling pathway(GO:0034121) negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.8 | 2.3 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.6 | 5.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 10.7 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.6 | 1.8 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.5 | 2.2 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.5 | 2.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.5 | 7.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.5 | 1.4 | GO:0007567 | parturition(GO:0007567) maternal process involved in parturition(GO:0060137) |
| 0.4 | 1.8 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.4 | 9.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.4 | 1.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 3.9 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 5.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.3 | 1.8 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.3 | 1.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.3 | 5.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.3 | 0.9 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.3 | 2.9 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 1.5 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 0.7 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 2.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 2.4 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 2.4 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.2 | 0.8 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 5.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 0.5 | GO:1900274 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.2 | 2.7 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.2 | 0.8 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.2 | 5.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.2 | 1.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 2.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.6 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 7.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 2.6 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 3.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 4.0 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 1.4 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 2.6 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 0.8 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 0.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 3.9 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 2.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 2.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 1.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 2.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 2.2 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 1.8 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 4.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 2.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 9.0 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 2.7 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 7.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 6.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 0.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 1.0 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 4.6 | GO:0016571 | histone methylation(GO:0016571) |
| 0.1 | 1.9 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.1 | 1.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.5 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 0.7 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.3 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 6.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 7.8 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 2.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 5.1 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 3.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 8.3 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 8.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.2 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.4 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 3.0 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.3 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 1.6 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 2.1 | GO:0043410 | positive regulation of MAPK cascade(GO:0043410) |
| 0.0 | 7.3 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 2.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.1 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 7.2 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 3.8 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 3.3 | GO:0032956 | regulation of actin cytoskeleton organization(GO:0032956) |
| 0.0 | 0.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.7 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.9 | 13.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.8 | 3.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.8 | 6.5 | GO:0042613 | MHC protein complex(GO:0042611) MHC class II protein complex(GO:0042613) |
| 0.6 | 9.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.4 | 6.2 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.4 | 5.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.4 | 2.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 5.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 2.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 7.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 0.8 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 6.1 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 15.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 7.5 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.1 | 4.0 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 8.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 2.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 16.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 3.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 10.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 2.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 7.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 71.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 7.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.7 | GO:0000793 | condensed chromosome(GO:0000793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 15.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.8 | 12.6 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 1.8 | 7.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.1 | 10.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.9 | 6.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.8 | 3.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.7 | 32.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.5 | 1.8 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.5 | 6.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.5 | 1.4 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.4 | 2.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 2.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.4 | 13.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.4 | 8.9 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.3 | 2.7 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.3 | 5.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 9.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 9.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 5.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 2.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.3 | 1.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 6.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.3 | 2.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 3.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 0.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.3 | 1.3 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.3 | 5.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 3.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.7 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.2 | 7.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 1.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 4.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 4.1 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 2.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 6.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.8 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 2.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 3.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 3.9 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 17.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 3.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 1.0 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.8 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 7.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.1 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 9.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 5.0 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.1 | 4.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.7 | GO:0002039 | p53 binding(GO:0002039) lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 3.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 5.0 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 4.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 6.1 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 4.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.4 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 4.4 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 6.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.1 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 7.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.8 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 2.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 9.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.1 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.5 | 4.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 16.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.4 | 9.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.4 | 10.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.3 | 3.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 8.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.3 | 4.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 4.8 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 5.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 7.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 2.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 4.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.5 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 15.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 0.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.1 | 7.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.5 | 3.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.4 | 5.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 6.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 4.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 2.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.3 | 10.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.2 | 4.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 4.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 5.2 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.1 | 4.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 1.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.0 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 2.1 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 1.5 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 2.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |