Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
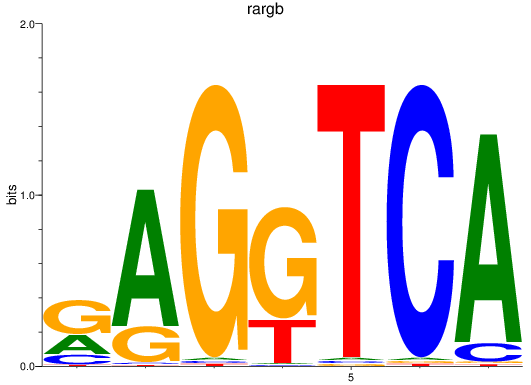
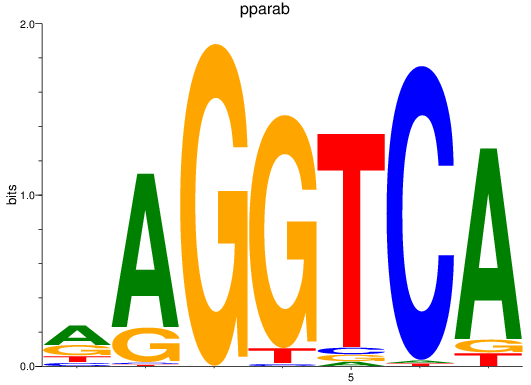
Results for rargb_pparab
Z-value: 1.94
Transcription factors associated with rargb_pparab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rargb
|
ENSDARG00000054003 | retinoic acid receptor, gamma b |
|
pparab
|
ENSDARG00000054323 | peroxisome proliferator-activated receptor alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pparab | dr11_v1_chr25_+_2198781_2198781 | -0.28 | 5.5e-03 | Click! |
| rargb | dr11_v1_chr11_+_2089461_2089461 | -0.02 | 8.2e-01 | Click! |
Activity profile of rargb_pparab motif
Sorted Z-values of rargb_pparab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_24289641 | 35.32 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr3_+_39568290 | 33.75 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr11_+_11201096 | 31.14 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr13_+_22480496 | 25.09 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr3_+_39566999 | 23.34 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr6_-_40722200 | 21.29 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr17_-_5583345 | 19.99 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr3_+_33300522 | 19.55 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr7_+_31871830 | 19.33 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr24_+_25467465 | 19.19 |
ENSDART00000189933
|
smpx
|
small muscle protein, X-linked |
| chr15_+_24644016 | 19.19 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr12_-_26415499 | 18.79 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr8_+_48613040 | 18.16 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr3_+_30257582 | 17.03 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr16_+_29650698 | 16.90 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr15_+_20403903 | 15.96 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr13_+_22480857 | 15.64 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr11_+_36243774 | 15.09 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr11_-_25213651 | 14.51 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr2_+_38039857 | 14.01 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr23_-_35756115 | 13.81 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr8_-_18535822 | 13.75 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr1_-_55810730 | 13.61 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr9_-_42861080 | 13.61 |
ENSDART00000193688
|
ttn.1
|
titin, tandem duplicate 1 |
| chr2_+_42191592 | 13.58 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr12_-_4683325 | 12.77 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr6_-_40722480 | 12.29 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr19_-_47555956 | 12.24 |
ENSDART00000114549
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr15_-_20933574 | 12.14 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr22_-_7050 | 11.32 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr6_-_15653494 | 11.05 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr10_+_22782522 | 11.03 |
ENSDART00000079498
ENSDART00000145558 |
si:ch211-237l4.6
|
si:ch211-237l4.6 |
| chr20_-_29483514 | 10.78 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr6_-_1514767 | 10.70 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr8_+_50534948 | 10.69 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr23_-_45705525 | 10.45 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr19_+_712127 | 10.43 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr18_+_8340886 | 10.03 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr11_+_42587900 | 9.97 |
ENSDART00000167529
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr23_+_42810055 | 9.81 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr11_-_11471857 | 9.72 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr21_+_5080789 | 9.65 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr5_-_68916455 | 9.52 |
ENSDART00000171465
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr3_-_32170850 | 9.48 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr10_+_6318227 | 9.45 |
ENSDART00000170872
ENSDART00000162428 ENSDART00000158994 |
tpm2
|
tropomyosin 2 (beta) |
| chr19_-_32783373 | 9.44 |
ENSDART00000145790
|
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr22_+_10201826 | 9.37 |
ENSDART00000006513
ENSDART00000132641 |
pdhb
|
pyruvate dehydrogenase E1 beta subunit |
| chr21_+_15704556 | 9.36 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr6_-_39764995 | 9.26 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr6_-_10912424 | 9.24 |
ENSDART00000036456
|
cycsb
|
cytochrome c, somatic b |
| chr25_+_3306620 | 9.11 |
ENSDART00000182085
ENSDART00000034704 |
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr6_-_10835849 | 9.11 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr15_+_24644251 | 9.03 |
ENSDART00000181660
|
smtnl
|
smoothelin, like |
| chr3_+_24207243 | 8.91 |
ENSDART00000023454
ENSDART00000136400 |
adsl
|
adenylosuccinate lyase |
| chr1_-_58868306 | 8.68 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr25_+_29160102 | 8.55 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr18_+_27926839 | 8.53 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr11_+_24313931 | 8.49 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr11_+_24314148 | 8.47 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr20_+_40150612 | 8.44 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr11_+_7158723 | 8.34 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr10_+_22775253 | 8.32 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr4_-_25836684 | 8.27 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr8_-_14184423 | 8.12 |
ENSDART00000063817
|
ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
| chr16_-_17197546 | 8.07 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr12_-_4070058 | 8.04 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr25_+_25124684 | 8.02 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr12_-_47648538 | 7.92 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr22_-_15593824 | 7.89 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr4_+_6572364 | 7.87 |
ENSDART00000122574
|
ppp1r3aa
|
protein phosphatase 1, regulatory subunit 3Aa |
| chr16_-_560574 | 7.85 |
ENSDART00000148452
|
irx2a
|
iroquois homeobox 2a |
| chr10_-_44017642 | 7.85 |
ENSDART00000135240
ENSDART00000014669 |
acads
|
acyl-CoA dehydrogenase short chain |
| chr6_-_49898881 | 7.83 |
ENSDART00000150204
|
atp5f1e
|
ATP synthase F1 subunit epsilon |
| chr8_-_17997845 | 7.78 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr15_+_404891 | 7.77 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr18_-_226800 | 7.74 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr2_-_44199722 | 7.69 |
ENSDART00000140633
ENSDART00000145728 |
sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr23_+_18722915 | 7.67 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr6_-_39765546 | 7.65 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr15_-_23645810 | 7.65 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr9_-_49493305 | 7.65 |
ENSDART00000148707
ENSDART00000148561 |
xirp2b
|
xin actin binding repeat containing 2b |
| chr9_+_54644626 | 7.65 |
ENSDART00000190609
|
egfl6
|
EGF-like-domain, multiple 6 |
| chr13_+_281214 | 7.48 |
ENSDART00000137572
|
mpc1
|
mitochondrial pyruvate carrier 1 |
| chr9_-_22821901 | 7.42 |
ENSDART00000101711
|
neb
|
nebulin |
| chr10_-_39153959 | 7.36 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr14_-_34771371 | 7.33 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr9_-_710896 | 7.22 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr20_+_54738210 | 7.20 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr23_+_5524247 | 7.20 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr19_-_32487469 | 7.06 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr3_-_16010968 | 6.94 |
ENSDART00000080672
|
mrps34
|
mitochondrial ribosomal protein S34 |
| chr18_-_16791331 | 6.93 |
ENSDART00000148222
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr4_+_38344 | 6.91 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr14_-_29799993 | 6.86 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr9_+_2333927 | 6.60 |
ENSDART00000123340
|
atp5mc3a
|
ATP synthase membrane subunit c locus 3a |
| chr16_+_30961822 | 6.58 |
ENSDART00000059187
|
gstk2
|
glutathione S-transferase kappa 2 |
| chr13_-_37254777 | 6.55 |
ENSDART00000139734
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr14_+_30340251 | 6.49 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr8_+_14158021 | 6.44 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr18_+_36037223 | 6.42 |
ENSDART00000144410
|
tmem91
|
transmembrane protein 91 |
| chr9_-_34945566 | 6.41 |
ENSDART00000131908
ENSDART00000059861 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr25_-_31396479 | 6.37 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr12_-_29305533 | 6.36 |
ENSDART00000189410
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr19_+_41169996 | 6.34 |
ENSDART00000048438
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr6_-_49873020 | 6.30 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr23_+_18722715 | 6.30 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr6_+_41463786 | 6.25 |
ENSDART00000065006
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr7_-_22941472 | 5.98 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr3_-_18410968 | 5.97 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr8_-_17980317 | 5.97 |
ENSDART00000129148
|
tnni3k
|
TNNI3 interacting kinase |
| chr14_-_12253309 | 5.93 |
ENSDART00000115101
|
myot
|
myotilin |
| chr17_+_53424415 | 5.90 |
ENSDART00000157022
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr24_+_25471196 | 5.90 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr20_+_1996202 | 5.88 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr14_-_17121676 | 5.85 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr14_+_33413980 | 5.84 |
ENSDART00000052780
ENSDART00000124437 ENSDART00000173327 |
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| chr9_+_30464641 | 5.82 |
ENSDART00000128357
|
gja5a
|
gap junction protein, alpha 5a |
| chr3_-_12187245 | 5.80 |
ENSDART00000189553
ENSDART00000165131 |
srl
|
sarcalumenin |
| chr14_-_33454595 | 5.73 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr8_+_25733872 | 5.72 |
ENSDART00000156340
|
si:ch211-167b20.8
|
si:ch211-167b20.8 |
| chr9_-_23944470 | 5.72 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr22_-_26595027 | 5.71 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr12_-_34827477 | 5.66 |
ENSDART00000153026
|
NDUFAF8
|
si:dkey-21c1.6 |
| chr2_-_48753873 | 5.57 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr4_-_4932619 | 5.56 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr23_-_35069805 | 5.52 |
ENSDART00000087219
|
BX294434.1
|
|
| chr9_-_22822084 | 5.45 |
ENSDART00000142020
|
neb
|
nebulin |
| chr7_-_51793333 | 5.40 |
ENSDART00000180654
|
BX957362.5
|
|
| chr1_+_23783349 | 5.40 |
ENSDART00000007531
|
slit2
|
slit homolog 2 (Drosophila) |
| chr3_+_20001608 | 5.40 |
ENSDART00000137944
|
asb16
|
ankyrin repeat and SOCS box containing 16 |
| chr13_+_22249636 | 5.35 |
ENSDART00000108472
ENSDART00000173123 |
synpo2la
|
synaptopodin 2-like a |
| chr25_+_19008497 | 5.35 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr18_+_30508729 | 5.28 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr13_+_24853578 | 5.27 |
ENSDART00000145865
|
si:dkey-24f15.2
|
si:dkey-24f15.2 |
| chr2_-_15318786 | 5.22 |
ENSDART00000135851
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr22_-_15587360 | 5.20 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr10_-_39154594 | 5.18 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr24_-_23974559 | 5.18 |
ENSDART00000080510
ENSDART00000135242 |
ndufb4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4 |
| chr17_-_38778826 | 5.17 |
ENSDART00000168182
ENSDART00000124041 ENSDART00000136921 |
dglucy
|
D-glutamate cyclase |
| chr13_+_28821841 | 5.17 |
ENSDART00000179900
|
CU639469.1
|
|
| chr20_+_19066596 | 5.12 |
ENSDART00000130271
|
sox7
|
SRY (sex determining region Y)-box 7 |
| chr21_-_23035120 | 5.12 |
ENSDART00000016502
|
dlat
|
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate dehydrogenase complex) |
| chr12_-_29305220 | 5.02 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr12_-_20373058 | 5.00 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr22_+_19473804 | 4.93 |
ENSDART00000135085
ENSDART00000147673 ENSDART00000156818 |
si:dkey-78l4.7
|
si:dkey-78l4.7 |
| chr17_+_135590 | 4.86 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr22_+_19247255 | 4.79 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr7_+_65586624 | 4.77 |
ENSDART00000184344
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr16_+_32736588 | 4.77 |
ENSDART00000075191
ENSDART00000168358 |
zgc:172323
|
zgc:172323 |
| chr23_+_17417539 | 4.75 |
ENSDART00000182605
|
BX649300.2
|
|
| chr17_-_17764801 | 4.74 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr7_-_41851605 | 4.72 |
ENSDART00000142981
|
mylk3
|
myosin light chain kinase 3 |
| chr20_-_23254876 | 4.71 |
ENSDART00000141510
|
ociad1
|
OCIA domain containing 1 |
| chr4_-_16451375 | 4.68 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr5_-_69316142 | 4.67 |
ENSDART00000157238
ENSDART00000144570 |
smtnb
|
smoothelin b |
| chr13_+_50375800 | 4.63 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr18_+_30507839 | 4.62 |
ENSDART00000026866
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr4_+_2620751 | 4.62 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr9_-_21067971 | 4.59 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr8_+_2530065 | 4.54 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr19_+_9212031 | 4.52 |
ENSDART00000052930
|
ndufv1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1 |
| chr6_+_52804267 | 4.52 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr17_-_31058664 | 4.52 |
ENSDART00000126414
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr8_-_979735 | 4.49 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr21_-_20381481 | 4.44 |
ENSDART00000115236
|
atp5mea
|
ATP synthase membrane subunit ea |
| chr23_-_9919959 | 4.42 |
ENSDART00000127029
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr19_+_30990815 | 4.42 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr25_-_29134654 | 4.40 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr12_+_49125510 | 4.38 |
ENSDART00000185804
|
FO704607.1
|
|
| chr23_+_44611864 | 4.30 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr1_-_59232267 | 4.27 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr11_+_77526 | 4.24 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr11_-_7410537 | 4.24 |
ENSDART00000009859
|
adgrl4
|
adhesion G protein-coupled receptor L4 |
| chr12_-_47782623 | 4.23 |
ENSDART00000115742
|
selenou1b
|
selenoprotein U1b |
| chr1_-_9109699 | 4.22 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr1_+_580642 | 4.15 |
ENSDART00000147633
|
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr22_+_19266995 | 4.15 |
ENSDART00000133995
ENSDART00000144963 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr21_+_10866421 | 4.14 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr13_-_13751017 | 4.04 |
ENSDART00000180182
|
ky
|
kyphoscoliosis peptidase |
| chr15_+_21882419 | 4.01 |
ENSDART00000157216
|
si:dkey-103g5.4
|
si:dkey-103g5.4 |
| chr11_+_21076872 | 4.00 |
ENSDART00000155521
|
prelp
|
proline/arginine-rich end leucine-rich repeat protein |
| chr5_+_36439405 | 3.98 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr5_-_46980651 | 3.98 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr16_-_32006737 | 3.97 |
ENSDART00000184813
ENSDART00000179827 |
gstk4
|
glutathione S-transferase kappa 4 |
| chr23_-_27571667 | 3.94 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr9_-_43142636 | 3.89 |
ENSDART00000134349
ENSDART00000181835 |
ccdc141
|
coiled-coil domain containing 141 |
| chr25_-_13363286 | 3.88 |
ENSDART00000163735
ENSDART00000169119 |
ndrg4
|
NDRG family member 4 |
| chr6_-_15096556 | 3.83 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr23_+_42813415 | 3.81 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr8_+_15251448 | 3.78 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr2_-_44255537 | 3.78 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr6_+_28124393 | 3.77 |
ENSDART00000089195
|
gpr17
|
G protein-coupled receptor 17 |
| chr3_+_27770110 | 3.74 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr5_-_18007077 | 3.74 |
ENSDART00000129878
|
zdhhc8b
|
zinc finger, DHHC-type containing 8b |
| chr3_-_32590164 | 3.74 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr16_-_2818170 | 3.71 |
ENSDART00000081918
|
acot22
|
acyl-CoA thioesterase 22 |
| chr24_+_2470061 | 3.70 |
ENSDART00000140383
ENSDART00000191261 |
F13A1 (1 of many)
|
coagulation factor XIII A chain |
| chr18_+_48423973 | 3.69 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr12_-_28983584 | 3.68 |
ENSDART00000112374
|
zgc:171713
|
zgc:171713 |
| chr3_+_58167288 | 3.68 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rargb_pparab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.2 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 4.7 | 14.0 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 4.5 | 13.6 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 3.9 | 19.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 3.8 | 22.8 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 3.7 | 87.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 3.4 | 17.0 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 2.9 | 8.8 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 2.8 | 17.0 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 2.8 | 8.3 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 2.8 | 16.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 2.7 | 13.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 2.1 | 12.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 2.0 | 13.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 2.0 | 7.8 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.9 | 48.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 1.8 | 5.4 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 1.7 | 12.0 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 1.6 | 14.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.5 | 4.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 1.5 | 15.1 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 1.4 | 9.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 1.3 | 18.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 1.3 | 16.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.3 | 5.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 1.3 | 3.8 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 1.2 | 12.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.2 | 23.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 1.2 | 3.6 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 1.1 | 6.8 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 1.0 | 3.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.0 | 5.1 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 1.0 | 12.9 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 1.0 | 8.9 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 1.0 | 4.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.9 | 2.8 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.9 | 9.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.9 | 2.7 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.9 | 2.7 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.8 | 2.5 | GO:0034398 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.8 | 20.5 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.8 | 22.9 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.8 | 1.6 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.8 | 4.8 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.7 | 3.0 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.7 | 2.8 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.7 | 2.7 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.6 | 1.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.6 | 2.4 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.6 | 6.0 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.6 | 13.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.6 | 5.1 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.6 | 3.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.5 | 12.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.5 | 4.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.5 | 24.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.5 | 2.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.5 | 3.1 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.5 | 2.1 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.5 | 0.5 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.5 | 2.5 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.5 | 0.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.5 | 10.8 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.5 | 17.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.5 | 2.3 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.5 | 4.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.5 | 2.3 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.5 | 8.1 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.4 | 0.9 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.4 | 4.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.4 | 1.7 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.4 | 7.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.4 | 2.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.4 | 2.5 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.4 | 2.9 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.4 | 2.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 2.4 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.4 | 1.2 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.4 | 1.6 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.4 | 9.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.4 | 2.7 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.4 | 1.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.4 | 2.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 5.7 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.4 | 1.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.4 | 1.4 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.4 | 5.3 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.4 | 1.8 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.4 | 15.1 | GO:0060840 | artery development(GO:0060840) |
| 0.3 | 10.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.3 | 1.4 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 1.4 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 0.3 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.3 | 61.6 | GO:0060538 | skeletal muscle organ development(GO:0060538) |
| 0.3 | 2.3 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 1.3 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.3 | 3.0 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.3 | 6.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 1.0 | GO:0035992 | tendon formation(GO:0035992) |
| 0.3 | 1.6 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) regulation of mitophagy(GO:1903146) |
| 0.3 | 0.3 | GO:0086001 | cardiac muscle cell action potential(GO:0086001) |
| 0.3 | 4.4 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.3 | 1.2 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.3 | 1.9 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.3 | 44.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.3 | 9.7 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.3 | 1.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.6 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.3 | 1.1 | GO:0021800 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.3 | 3.9 | GO:0060038 | striated muscle cell proliferation(GO:0014855) cardiac muscle cell proliferation(GO:0060038) |
| 0.3 | 0.8 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.3 | 0.5 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.3 | 6.7 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.3 | 1.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 3.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 1.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 2.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 1.2 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.7 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.2 | 7.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 1.2 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.2 | 0.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.2 | 0.9 | GO:0090148 | membrane fission(GO:0090148) |
| 0.2 | 1.8 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 1.4 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 1.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 1.6 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) |
| 0.2 | 30.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.2 | 1.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 4.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 1.3 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 1.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.2 | 2.2 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.2 | 1.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 2.2 | GO:0098659 | inorganic cation import into cell(GO:0098659) inorganic ion import into cell(GO:0099587) |
| 0.2 | 1.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 4.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.2 | 0.4 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.2 | 1.7 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 1.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 2.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 0.6 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 1.2 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 0.8 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.2 | 0.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 0.8 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.2 | 1.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.6 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.2 | 1.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 0.8 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 0.8 | GO:0048331 | axial mesoderm formation(GO:0048320) axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) axial mesoderm structural organization(GO:0048331) mesoderm structural organization(GO:0048338) |
| 0.2 | 0.8 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.2 | 2.1 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 1.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 2.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 5.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.2 | 0.7 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 1.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 1.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.2 | 3.1 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.2 | 1.3 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.2 | 0.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 2.4 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 3.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 0.7 | GO:0033605 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.2 | 0.5 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.2 | 0.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 1.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.0 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 0.9 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 0.5 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.2 | 1.2 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.2 | 2.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 0.5 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 1.0 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.2 | 4.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.2 | 0.3 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.2 | 2.4 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.2 | 1.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 5.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 | 4.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 1.2 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.1 | 0.7 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.1 | 1.9 | GO:0046545 | development of primary female sexual characteristics(GO:0046545) |
| 0.1 | 2.8 | GO:0030168 | platelet activation(GO:0030168) |
| 0.1 | 0.8 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 0.6 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 7.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 2.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 1.0 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 5.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 11.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.4 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.9 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.6 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 2.5 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.6 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 7.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 4.4 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 2.8 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.1 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.6 | GO:0071265 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 1.7 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 4.7 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.1 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.1 | 1.0 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.8 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 1.7 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 2.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 1.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 1.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 2.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 0.8 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 2.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 7.1 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 0.4 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 2.2 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.1 | 1.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 7.5 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.1 | 2.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.1 | 0.8 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 3.6 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.1 | 0.5 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body catabolic process(GO:0046952) ketone body metabolic process(GO:1902224) |
| 0.1 | 1.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.6 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 3.0 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.4 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 3.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.2 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.7 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.4 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 1.5 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 3.0 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 5.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 1.4 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.8 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 1.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.6 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.4 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.7 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 2.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 8.8 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.1 | 1.2 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.9 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 1.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.2 | GO:0005991 | disaccharide metabolic process(GO:0005984) trehalose metabolic process(GO:0005991) |
| 0.1 | 1.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.8 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 2.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.4 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.9 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.8 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 1.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 4.4 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.1 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.9 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 1.3 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.1 | 1.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.1 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.7 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.4 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 2.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.2 | GO:0006867 | asparagine transport(GO:0006867) |
| 0.1 | 19.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 2.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.7 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.4 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.4 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 5.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 1.5 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 4.5 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 4.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.6 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 1.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.9 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 2.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.7 | GO:1901386 | negative regulation of calcium ion transmembrane transporter activity(GO:1901020) negative regulation of voltage-gated calcium channel activity(GO:1901386) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 2.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 4.6 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.2 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.0 | 0.5 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.6 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.0 | 0.2 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.0 | 0.2 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.0 | 1.3 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.8 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.7 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.6 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 4.5 | GO:0007005 | mitochondrion organization(GO:0007005) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 1.6 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 2.1 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 2.0 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.4 | GO:0016444 | somatic cell DNA recombination(GO:0016444) |
| 0.0 | 0.0 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.7 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.6 | GO:0046503 | glycerolipid catabolic process(GO:0046503) |
| 0.0 | 1.3 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.8 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.7 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.9 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 3.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.0 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 1.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.2 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.8 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 1.0 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.8 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 12.2 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 2.0 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.7 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.6 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0045823 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) positive regulation of heart contraction(GO:0045823) |
| 0.0 | 0.2 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0050921 | positive regulation of chemotaxis(GO:0050921) regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.7 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0072073 | nephron epithelium development(GO:0072009) kidney epithelium development(GO:0072073) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 25.6 | GO:0043034 | costamere(GO:0043034) |
| 3.4 | 13.6 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 3.3 | 23.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 2.8 | 17.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 2.8 | 13.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 2.4 | 7.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 2.0 | 9.9 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.9 | 20.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.9 | 13.0 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 1.7 | 25.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.7 | 3.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.6 | 31.1 | GO:0031430 | M band(GO:0031430) |
| 1.5 | 62.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 1.5 | 40.8 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 1.4 | 23.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.3 | 5.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 1.3 | 58.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 1.2 | 12.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.1 | 8.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.9 | 2.6 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.8 | 13.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.8 | 16.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.8 | 11.3 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.8 | 43.7 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.7 | 2.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.7 | 1.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.7 | 53.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.6 | 2.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 9.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.5 | 4.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 21.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.5 | 6.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.5 | 4.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.5 | 60.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.5 | 2.9 | GO:0000801 | central element(GO:0000801) |
| 0.5 | 1.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.4 | 9.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 2.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.4 | 7.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.4 | 2.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 2.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 24.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.4 | 12.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.4 | 3.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 8.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 14.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.3 | 3.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.3 | 4.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 15.5 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.3 | 4.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 7.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.3 | 10.5 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 2.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 1.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.3 | 1.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 1.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.7 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 5.2 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 1.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.8 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 0.7 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.2 | 1.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 0.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 1.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 0.6 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.2 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 0.9 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.2 | 2.5 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 14.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 22.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 1.5 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 10.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 1.5 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 7.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.0 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.0 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 1.7 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.1 | 3.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 2.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.5 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 3.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 7.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 6.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.1 | GO:0032421 | stereocilium bundle(GO:0032421) |
| 0.1 | 4.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 5.7 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 11.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 1.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 9.2 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.1 | 2.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.4 | GO:0034704 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.1 | 1.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 1.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 3.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 2.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.8 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.8 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 3.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 38.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 16.3 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 2.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.1 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 22.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.4 | 66.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 3.3 | 9.9 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 2.6 | 12.8 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 2.4 | 17.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 2.4 | 47.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 2.4 | 7.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 2.2 | 8.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 2.1 | 23.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 2.0 | 8.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.9 | 20.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.8 | 19.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.6 | 10.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 1.5 | 6.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 1.4 | 8.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.4 | 30.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 1.3 | 3.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.3 | 8.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 1.3 | 5.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 1.3 | 6.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.2 | 9.7 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 1.2 | 4.8 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 1.2 | 33.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 1.1 | 6.8 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 1.1 | 3.2 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 1.0 | 22.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.0 | 8.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.9 | 13.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.9 | 2.6 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.8 | 19.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.7 | 11.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.7 | 14.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.7 | 3.5 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.7 | 4.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.7 | 3.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.7 | 9.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.6 | 3.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.6 | 20.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.6 | 3.0 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.6 | 2.9 | GO:0015288 | porin activity(GO:0015288) |
| 0.6 | 4.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 1.6 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.5 | 2.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 4.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 3.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.5 | 1.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.5 | 1.4 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.5 | 4.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 2.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.5 | 2.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.5 | 6.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.4 | 4.0 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 2.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 1.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.4 | 1.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 0.9 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.4 | 3.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 2.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 2.4 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.4 | 153.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.4 | 1.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 4.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 1.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.4 | 1.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 3.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 1.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.4 | 1.4 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.4 | 2.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.4 | 4.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 1.4 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 5.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 24.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.3 | 2.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 1.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.3 | 4.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 17.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 1.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.3 | 5.7 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.3 | 2.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.3 | 25.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.3 | 1.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 9.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 2.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 1.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 4.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.3 | 2.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 1.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.3 | 8.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 3.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 15.4 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.3 | 4.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 0.7 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.2 | 1.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 2.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.2 | 0.7 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.2 | 1.3 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 6.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 1.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 3.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 64.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.2 | 5.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 4.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.2 | 1.6 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.2 | 1.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 1.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 1.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.4 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.2 | 3.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 1.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 7.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.2 | 3.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 2.4 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 0.7 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.2 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 1.3 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.2 | 1.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 2.0 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 3.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.2 | 8.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 2.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 8.2 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.2 | 2.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 2.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.6 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 0.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 7.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.0 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 1.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.6 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 4.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 2.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 24.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.8 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 9.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.9 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 2.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.4 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.1 | 0.7 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 2.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 2.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.1 | 1.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.7 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.8 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.4 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 0.7 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 5.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 0.7 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 1.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.7 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.7 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.8 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 1.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) excitatory extracellular ligand-gated ion channel activity(GO:0005231) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.6 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.4 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 1.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.5 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 2.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.8 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 3.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.2 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 0.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 1.6 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.2 | GO:0004555 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.1 | 0.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.1 | 1.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 0.2 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.1 | 1.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 1.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 3.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 1.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 1.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.5 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 7.9 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.7 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 9.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 3.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.2 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.1 | 1.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 2.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.2 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 2.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.4 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.0 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 1.2 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 1.4 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 7.3 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 8.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.7 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) ion gated channel activity(GO:0022839) |
| 0.0 | 1.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 1.2 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 1.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 4.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 6.3 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 2.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 1.2 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 8.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 11.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.2 | GO:0005230 | extracellular ligand-gated ion channel activity(GO:0005230) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 13.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 14.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 15.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.3 | 6.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 2.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.2 | 5.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 2.4 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 5.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 12.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 5.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 97.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 1.2 | 11.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 1.2 | 14.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.1 | 15.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.1 | 31.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.0 | 14.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.9 | 8.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.8 | 12.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.8 | 15.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.8 | 11.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.8 | 2.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.7 | 7.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.6 | 8.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.6 | 12.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 8.6 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.5 | 3.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.4 | 4.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 2.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.3 | 2.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 4.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.3 | 2.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 2.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 3.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 2.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 9.6 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.2 | 2.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 7.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.2 | 1.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 2.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 2.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 0.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 17.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.2 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 1.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 3.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |