Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
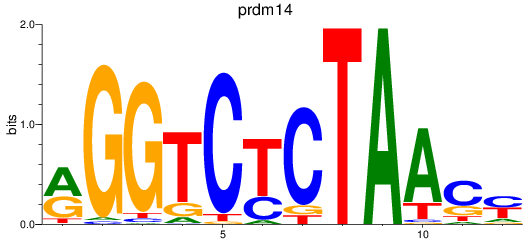
Results for prdm14
Z-value: 0.76
Transcription factors associated with prdm14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm14
|
ENSDARG00000045371 | PR domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm14 | dr11_v1_chr24_+_14451404_14451404 | 0.27 | 7.6e-03 | Click! |
Activity profile of prdm14 motif
Sorted Z-values of prdm14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_27020201 | 15.82 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr11_-_23080970 | 10.43 |
ENSDART00000127791
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr14_-_30642819 | 10.39 |
ENSDART00000078154
|
npas4a
|
neuronal PAS domain protein 4a |
| chr25_-_8030425 | 8.27 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr21_-_43949208 | 7.69 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr5_-_45877387 | 6.34 |
ENSDART00000183714
ENSDART00000041503 |
slc4a4a
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4a |
| chr10_-_43721530 | 5.74 |
ENSDART00000025366
|
cetn3
|
centrin 3 |
| chr21_-_21089781 | 5.71 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr11_-_18791834 | 5.65 |
ENSDART00000156431
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr4_-_22207873 | 5.62 |
ENSDART00000142140
|
ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr25_-_8030113 | 5.61 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr5_+_49744713 | 5.23 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr22_-_26323893 | 5.10 |
ENSDART00000105099
|
capn1b
|
calpain 1, (mu/I) large subunit b |
| chr22_-_20342260 | 4.91 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr8_-_25566347 | 4.74 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr22_-_11137268 | 4.40 |
ENSDART00000178882
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr1_+_41849152 | 4.39 |
ENSDART00000053685
|
smox
|
spermine oxidase |
| chr12_+_39685485 | 4.38 |
ENSDART00000163403
|
LO017650.1
|
|
| chr23_+_39558508 | 4.07 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr7_+_13418812 | 3.91 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr22_-_11136625 | 3.86 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr5_+_37903790 | 3.25 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr1_+_46509176 | 3.15 |
ENSDART00000166028
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr5_-_67115872 | 3.09 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr2_-_32826108 | 3.07 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr20_-_46467280 | 3.05 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr17_+_27162367 | 3.00 |
ENSDART00000193345
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr6_-_24384654 | 2.99 |
ENSDART00000164723
|
brdt
|
bromodomain, testis-specific |
| chr7_+_48806420 | 2.87 |
ENSDART00000083431
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr10_-_32877348 | 2.86 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr25_-_1235457 | 2.85 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr5_-_65081600 | 2.54 |
ENSDART00000160850
|
inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr16_+_32014552 | 2.50 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr8_-_25716074 | 2.46 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr21_-_14692119 | 2.46 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr25_+_16043246 | 2.40 |
ENSDART00000186663
|
sb:cb470
|
sb:cb470 |
| chr7_+_48805534 | 2.22 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr24_+_26402110 | 2.19 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr9_+_33261330 | 2.16 |
ENSDART00000135384
|
usp9
|
ubiquitin specific peptidase 9 |
| chr20_+_51061695 | 2.12 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr9_-_33477588 | 2.12 |
ENSDART00000144150
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr23_+_21261313 | 2.08 |
ENSDART00000104268
ENSDART00000159046 |
emc1
|
ER membrane protein complex subunit 1 |
| chr8_+_35172594 | 2.06 |
ENSDART00000177146
|
BX897670.1
|
|
| chr22_+_39074688 | 1.88 |
ENSDART00000153547
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr7_+_48805725 | 1.87 |
ENSDART00000166543
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr5_+_51833305 | 1.77 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr21_+_44300689 | 1.76 |
ENSDART00000186298
ENSDART00000142810 |
gabra3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr5_-_69482891 | 1.74 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr15_+_20363859 | 1.73 |
ENSDART00000166846
|
supt5h
|
SPT5 homolog, DSIF elongation factor subunit |
| chr5_+_26079178 | 1.70 |
ENSDART00000145920
|
si:dkey-201c13.2
|
si:dkey-201c13.2 |
| chr15_+_19293744 | 1.69 |
ENSDART00000184994
ENSDART00000123815 |
jam3a
|
junctional adhesion molecule 3a |
| chr5_+_28271412 | 1.63 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr7_+_51324834 | 1.35 |
ENSDART00000114429
|
usp12b
|
ubiquitin specific peptidase 12b |
| chr23_+_5104743 | 1.35 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr5_+_51833132 | 1.32 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr6_+_18142623 | 1.27 |
ENSDART00000169431
ENSDART00000158841 |
si:dkey-237i9.8
|
si:dkey-237i9.8 |
| chr17_+_25871304 | 1.27 |
ENSDART00000185143
|
wapla
|
WAPL cohesin release factor a |
| chr23_+_9522781 | 1.26 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr6_-_21534301 | 1.26 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr20_-_51727860 | 1.20 |
ENSDART00000147044
|
brox
|
BRO1 domain and CAAX motif containing |
| chr2_-_36819624 | 1.20 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr14_-_30897177 | 1.18 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr13_+_18321140 | 1.18 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr23_+_9522942 | 1.17 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr2_+_32826235 | 1.14 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr13_+_23157053 | 1.05 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr15_-_42760110 | 1.03 |
ENSDART00000152490
|
si:ch211-181d7.3
|
si:ch211-181d7.3 |
| chr16_-_24135508 | 0.95 |
ENSDART00000171819
ENSDART00000103176 |
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr21_-_27272657 | 0.93 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr12_-_6818676 | 0.90 |
ENSDART00000106391
|
pcdh15b
|
protocadherin-related 15b |
| chr23_-_27442544 | 0.89 |
ENSDART00000019521
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr15_-_21837207 | 0.81 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr2_-_37458527 | 0.80 |
ENSDART00000146820
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr13_+_49727333 | 0.79 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr4_+_30775376 | 0.73 |
ENSDART00000158528
|
si:dkey-11d20.1
|
si:dkey-11d20.1 |
| chr20_-_4049862 | 0.70 |
ENSDART00000158057
|
sprtn
|
SprT-like N-terminal domain |
| chr6_-_21988375 | 0.67 |
ENSDART00000161257
|
plxnb1b
|
plexin b1b |
| chr11_+_13642157 | 0.65 |
ENSDART00000060251
|
wdr18
|
WD repeat domain 18 |
| chr22_-_9890386 | 0.65 |
ENSDART00000132304
|
si:dkey-253d23.5
|
si:dkey-253d23.5 |
| chr7_+_13491452 | 0.59 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr9_-_34945566 | 0.55 |
ENSDART00000131908
ENSDART00000059861 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr17_+_28675120 | 0.54 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr11_+_31380495 | 0.53 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr19_-_4851411 | 0.40 |
ENSDART00000110398
|
fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr9_+_8364553 | 0.32 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr13_+_4871886 | 0.32 |
ENSDART00000132301
|
micu1
|
mitochondrial calcium uptake 1 |
| chr6_+_518979 | 0.27 |
ENSDART00000151012
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr17_-_42492668 | 0.15 |
ENSDART00000183946
|
CR925755.2
|
|
| chr5_+_62988916 | 0.13 |
ENSDART00000123243
|
TIMM22
|
translocase of inner mitochondrial membrane 22 |
| chr21_-_27273147 | 0.09 |
ENSDART00000143239
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 2.1 | 10.4 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 1.0 | 3.0 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 1.0 | 3.9 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.7 | 7.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.6 | 2.5 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.6 | 4.1 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.6 | 5.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.5 | 3.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.4 | 1.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.4 | 5.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 1.7 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.3 | 8.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 6.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 1.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.2 | 2.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.2 | 1.2 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 4.7 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 2.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 7.7 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.9 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 1.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.9 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 16.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 1.8 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 1.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 9.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.7 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 2.5 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.7 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 4.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 5.6 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 2.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 3.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.8 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 15.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.6 | 2.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.6 | 1.7 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.5 | 8.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 3.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 10.4 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 2.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 2.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.8 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.1 | 6.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 7.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 3.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 4.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 12.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 20.3 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 5.7 | GO:0005815 | microtubule organizing center(GO:0005815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.9 | 4.4 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.5 | 25.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.5 | 6.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 5.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 5.6 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 10.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 3.9 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 3.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.2 | 2.5 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.2 | 3.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 5.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 2.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.8 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 2.5 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 1.9 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 2.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 3.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 5.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 3.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 8.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 7.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 8.8 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 2.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.6 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 8.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 7.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 3.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.6 | 2.5 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.5 | 10.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 4.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 3.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.2 | 3.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 3.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 4.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.7 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 1.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |