Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
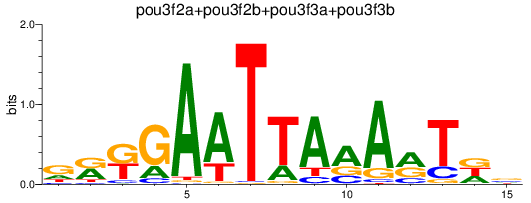
Results for pou3f2a+pou3f2b+pou3f3a+pou3f3b
Z-value: 1.32
Transcription factors associated with pou3f2a+pou3f2b+pou3f3a+pou3f3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou3f3a
|
ENSDARG00000042032 | POU class 3 homeobox 3a |
|
pou3f2a
|
ENSDARG00000070220 | POU class 3 homeobox 2a |
|
pou3f2b
|
ENSDARG00000076262 | POU class 3 homeobox 2b |
|
pou3f3b
|
ENSDARG00000095896 | POU class 3 homeobox 3b |
|
pou3f2b
|
ENSDARG00000112713 | POU class 3 homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou3f3a | dr11_v1_chr9_-_6661657_6661666 | -0.03 | 7.7e-01 | Click! |
| pou3f2b | dr11_v1_chr16_+_32559821_32559821 | 0.01 | 9.4e-01 | Click! |
| pou3f2a | dr11_v1_chr4_+_5741733_5741733 | -0.01 | 9.4e-01 | Click! |
| pou3f3b | dr11_v1_chr6_+_14949950_14949950 | 0.01 | 9.5e-01 | Click! |
Activity profile of pou3f2a+pou3f2b+pou3f3a+pou3f3b motif
Sorted Z-values of pou3f2a+pou3f2b+pou3f3a+pou3f3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_44945224 | 9.37 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr4_+_73085993 | 7.34 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr7_-_16562200 | 7.00 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr5_-_65037525 | 6.67 |
ENSDART00000158856
|
anxa1b
|
annexin A1b |
| chr16_-_24518027 | 6.32 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr23_+_36653376 | 6.16 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr3_-_32596394 | 6.14 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr24_-_26310854 | 5.98 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr5_-_65037371 | 5.61 |
ENSDART00000170560
|
anxa1b
|
annexin A1b |
| chr18_+_7591381 | 5.52 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr8_-_44378391 | 5.42 |
ENSDART00000158784
ENSDART00000098138 |
si:busm1-228j01.6
|
si:busm1-228j01.6 |
| chr7_+_3597413 | 5.36 |
ENSDART00000064281
|
si:dkey-192d15.2
|
si:dkey-192d15.2 |
| chr23_-_24263474 | 5.36 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr11_-_25418856 | 5.30 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr16_-_41990421 | 5.30 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr19_+_48290786 | 5.26 |
ENSDART00000165986
|
zgc:101785
|
zgc:101785 |
| chr3_+_49021079 | 5.24 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr16_+_23487051 | 5.22 |
ENSDART00000145496
|
icn2
|
ictacalcin 2 |
| chr15_-_29598679 | 5.19 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr5_-_55560937 | 5.12 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr19_-_5380770 | 4.97 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr9_+_30421489 | 4.97 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr20_-_29482492 | 4.76 |
ENSDART00000178308
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr22_-_24297510 | 4.69 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr18_-_12295092 | 4.66 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr3_+_55093581 | 4.44 |
ENSDART00000101709
|
hbaa2
|
hemoglobin, alpha adult 2 |
| chr16_+_29516098 | 4.42 |
ENSDART00000174895
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr6_+_584632 | 4.31 |
ENSDART00000151150
|
zgc:92360
|
zgc:92360 |
| chr24_+_3963684 | 4.24 |
ENSDART00000182959
ENSDART00000185926 ENSDART00000167043 ENSDART00000033394 |
pfkpa
|
phosphofructokinase, platelet a |
| chr10_+_35347993 | 4.10 |
ENSDART00000131350
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr22_-_31020690 | 4.06 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr20_+_28364742 | 4.04 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr11_+_25064519 | 3.95 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr16_+_35916371 | 3.93 |
ENSDART00000167208
|
sh3d21
|
SH3 domain containing 21 |
| chr2_+_36121373 | 3.91 |
ENSDART00000187002
|
CT867973.2
|
|
| chr8_+_21353878 | 3.86 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr7_+_20109968 | 3.77 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr8_+_4697764 | 3.76 |
ENSDART00000064197
|
cldn5a
|
claudin 5a |
| chr13_-_36761379 | 3.66 |
ENSDART00000131534
ENSDART00000029824 |
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr21_+_11248448 | 3.61 |
ENSDART00000142431
|
arid6
|
AT-rich interaction domain 6 |
| chr3_-_16537077 | 3.60 |
ENSDART00000133907
|
eps8l1
|
eps8-like1 |
| chr3_+_2971852 | 3.59 |
ENSDART00000059271
|
BX004816.1
|
|
| chr5_+_37091626 | 3.57 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr8_-_32497581 | 3.55 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr7_+_66822229 | 3.55 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr17_-_50010121 | 3.54 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr21_-_42831033 | 3.52 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr16_-_38001040 | 3.50 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr24_+_25471196 | 3.50 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr14_-_14640401 | 3.47 |
ENSDART00000168027
ENSDART00000167521 |
znf185
|
zinc finger protein 185 with LIM domain |
| chr5_-_33259079 | 3.44 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr16_+_21790870 | 3.39 |
ENSDART00000155039
|
trim108
|
tripartite motif containing 108 |
| chr8_-_36140405 | 3.39 |
ENSDART00000182806
|
CT583723.2
|
|
| chr19_+_2631565 | 3.29 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr25_+_15647750 | 3.21 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr12_-_46228023 | 3.21 |
ENSDART00000153455
|
si:ch211-226h7.6
|
si:ch211-226h7.6 |
| chr25_+_5035343 | 3.20 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr4_-_67799941 | 3.13 |
ENSDART00000185830
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr8_+_36416285 | 3.12 |
ENSDART00000158400
|
si:busm1-194e12.8
|
si:busm1-194e12.8 |
| chr13_-_33362191 | 3.10 |
ENSDART00000100514
|
zgc:172120
|
zgc:172120 |
| chr9_+_24008879 | 3.09 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr8_+_32402441 | 3.08 |
ENSDART00000191451
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr6_-_15101477 | 3.07 |
ENSDART00000187713
ENSDART00000124132 |
fhl2b
|
four and a half LIM domains 2b |
| chr23_-_37546696 | 3.07 |
ENSDART00000126892
|
tor1l2
|
torsin family 1 like 2 |
| chr16_-_32221312 | 3.05 |
ENSDART00000112348
|
calhm5.1
|
calcium homeostasis modulator family member 5, tandem duplicate 1 |
| chr16_+_10776688 | 3.04 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr11_+_4026229 | 3.04 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr21_-_26089964 | 3.03 |
ENSDART00000027848
|
tlcd1
|
TLC domain containing 1 |
| chr7_+_48761875 | 3.02 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr4_-_9214288 | 2.99 |
ENSDART00000067309
|
avpr1ab
|
arginine vasopressin receptor 1Ab |
| chr1_+_45056371 | 2.97 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr10_-_42237304 | 2.96 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr18_-_12327426 | 2.93 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr3_-_43770876 | 2.93 |
ENSDART00000160162
|
zgc:92162
|
zgc:92162 |
| chr15_-_35960250 | 2.91 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr17_-_15382704 | 2.89 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr5_+_29794058 | 2.88 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr15_+_38018301 | 2.85 |
ENSDART00000114188
ENSDART00000171150 |
si:ch73-380l3.1
|
si:ch73-380l3.1 |
| chr23_+_26142613 | 2.84 |
ENSDART00000165046
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr8_-_36469117 | 2.79 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr19_+_22850657 | 2.79 |
ENSDART00000130472
|
pkhd1l1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 |
| chr2_-_14656669 | 2.78 |
ENSDART00000190219
|
CABZ01026681.1
|
|
| chr19_-_5769728 | 2.78 |
ENSDART00000133106
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr13_-_22766445 | 2.78 |
ENSDART00000140151
|
si:ch211-150i13.1
|
si:ch211-150i13.1 |
| chr8_-_31107537 | 2.76 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr7_+_32901658 | 2.74 |
ENSDART00000115420
|
ano9b
|
anoctamin 9b |
| chr8_-_25074319 | 2.73 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr20_+_36806398 | 2.70 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr9_-_11551608 | 2.70 |
ENSDART00000128629
|
fev
|
FEV (ETS oncogene family) |
| chr25_-_37084032 | 2.70 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr16_+_17715243 | 2.70 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr6_+_2048920 | 2.69 |
ENSDART00000153568
|
quo
|
quattro |
| chr1_+_44906517 | 2.67 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr14_+_15597049 | 2.65 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr10_+_8527196 | 2.65 |
ENSDART00000141147
|
si:ch211-193e13.5
|
si:ch211-193e13.5 |
| chr18_-_46258612 | 2.60 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr2_+_10280645 | 2.59 |
ENSDART00000063996
|
gadd45aa
|
growth arrest and DNA-damage-inducible, alpha, a |
| chr18_-_7539166 | 2.56 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr10_+_33588715 | 2.56 |
ENSDART00000051198
|
mis18a
|
MIS18 kinetochore protein A |
| chr15_-_20190052 | 2.56 |
ENSDART00000157149
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr18_-_48296793 | 2.53 |
ENSDART00000032184
ENSDART00000193076 |
CABZ01069595.1
|
|
| chr14_+_36889893 | 2.52 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr5_-_30715225 | 2.50 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr17_-_14701529 | 2.49 |
ENSDART00000185142
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr7_-_38658411 | 2.48 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr7_-_19923249 | 2.48 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr10_+_40792078 | 2.45 |
ENSDART00000140007
|
si:ch211-139n6.3
|
si:ch211-139n6.3 |
| chr16_+_23796612 | 2.43 |
ENSDART00000131698
|
rab13
|
RAB13, member RAS oncogene family |
| chr1_-_10640748 | 2.42 |
ENSDART00000103550
|
zgc:174945
|
zgc:174945 |
| chr24_+_20920563 | 2.42 |
ENSDART00000037224
|
cst14a.2
|
cystatin 14a, tandem duplicate 2 |
| chr4_+_18843015 | 2.40 |
ENSDART00000152086
ENSDART00000066977 ENSDART00000132567 |
bik
|
BCL2 interacting killer |
| chr6_-_19270484 | 2.40 |
ENSDART00000186894
ENSDART00000188709 |
zgc:174863
|
zgc:174863 |
| chr22_+_789795 | 2.39 |
ENSDART00000185230
ENSDART00000192538 ENSDART00000171045 ENSDART00000180188 |
cry1bb
|
cryptochrome circadian clock 1bb |
| chr15_+_29116063 | 2.38 |
ENSDART00000016112
ENSDART00000153609 ENSDART00000155630 |
capns1b
|
calpain, small subunit 1 b |
| chr15_+_963292 | 2.38 |
ENSDART00000156586
|
alox5b.2
|
arachidonate 5-lipoxygenase b, tandem duplicate 2 |
| chr5_+_51026563 | 2.38 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr3_-_26921475 | 2.37 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr16_+_10777116 | 2.36 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr5_+_62611400 | 2.33 |
ENSDART00000132054
|
abr
|
active BCR-related |
| chr2_+_48074243 | 2.32 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr4_-_16412084 | 2.32 |
ENSDART00000188460
|
dcn
|
decorin |
| chr3_+_16841942 | 2.32 |
ENSDART00000023985
ENSDART00000145317 |
stk17al
|
serine/threonine kinase 17a like |
| chr10_+_22527715 | 2.30 |
ENSDART00000134864
|
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr19_-_5769553 | 2.29 |
ENSDART00000175003
|
si:ch211-264f5.6
|
si:ch211-264f5.6 |
| chr1_-_58000438 | 2.28 |
ENSDART00000163761
|
si:ch211-114l13.9
|
si:ch211-114l13.9 |
| chr7_-_41851605 | 2.28 |
ENSDART00000142981
|
mylk3
|
myosin light chain kinase 3 |
| chr16_-_26140768 | 2.27 |
ENSDART00000143960
|
cd79a
|
CD79a molecule, immunoglobulin-associated alpha |
| chr10_+_42542517 | 2.26 |
ENSDART00000005496
|
kctd9b
|
potassium channel tetramerization domain containing 9b |
| chr4_-_32642681 | 2.24 |
ENSDART00000188695
ENSDART00000193974 |
si:dkey-265e7.1
|
si:dkey-265e7.1 |
| chr11_+_39928828 | 2.24 |
ENSDART00000137516
ENSDART00000134082 |
vamp3
|
vesicle-associated membrane protein 3 (cellubrevin) |
| chr2_-_55337585 | 2.23 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr4_+_71578099 | 2.23 |
ENSDART00000180472
ENSDART00000162184 |
si:dkey-27n6.4
|
si:dkey-27n6.4 |
| chr19_-_15229421 | 2.22 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr21_-_41305748 | 2.22 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr10_+_36662640 | 2.21 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr23_-_31436524 | 2.21 |
ENSDART00000140519
|
zgc:153284
|
zgc:153284 |
| chr23_-_31372639 | 2.20 |
ENSDART00000179908
ENSDART00000135620 ENSDART00000053367 |
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr21_-_38853737 | 2.19 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr13_+_12405051 | 2.19 |
ENSDART00000108543
|
atp10d
|
ATPase phospholipid transporting 10D |
| chr16_+_27543893 | 2.19 |
ENSDART00000182421
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
| chr21_-_44565862 | 2.19 |
ENSDART00000180158
|
fundc2
|
fun14 domain containing 2 |
| chr22_+_8612462 | 2.16 |
ENSDART00000114586
|
CR450686.1
|
|
| chr7_-_8156169 | 2.16 |
ENSDART00000182960
ENSDART00000190184 ENSDART00000190885 ENSDART00000189867 ENSDART00000160836 |
si:cabz01030277.1
si:ch211-163c2.3
|
si:cabz01030277.1 si:ch211-163c2.3 |
| chr18_-_48983690 | 2.16 |
ENSDART00000182359
|
FO681288.3
|
|
| chr6_-_8480815 | 2.16 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr10_-_5684326 | 2.14 |
ENSDART00000157776
|
st8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr17_-_51893123 | 2.13 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr23_+_35847200 | 2.12 |
ENSDART00000129222
|
rarga
|
retinoic acid receptor gamma a |
| chr17_-_45150763 | 2.12 |
ENSDART00000155043
ENSDART00000156786 ENSDART00000191147 |
tmed8
|
transmembrane p24 trafficking protein 8 |
| chr21_-_22892124 | 2.10 |
ENSDART00000065563
|
ccdc90b
|
coiled-coil domain containing 90B |
| chr10_+_35275965 | 2.09 |
ENSDART00000077404
|
pora
|
P450 (cytochrome) oxidoreductase a |
| chr7_+_21752168 | 2.09 |
ENSDART00000173641
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr15_+_3808996 | 2.07 |
ENSDART00000110227
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr18_-_46208581 | 2.06 |
ENSDART00000141278
|
si:ch211-14c7.2
|
si:ch211-14c7.2 |
| chr16_-_31525290 | 2.06 |
ENSDART00000109996
|
wisp1b
|
WNT1 inducible signaling pathway protein 1b |
| chr11_-_21585176 | 2.05 |
ENSDART00000045391
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr4_+_20063279 | 2.05 |
ENSDART00000024925
|
gcc1
|
GRIP and coiled-coil domain containing 1 |
| chr5_+_30384554 | 2.05 |
ENSDART00000135483
|
zgc:158412
|
zgc:158412 |
| chr9_-_31108285 | 2.04 |
ENSDART00000003193
|
gpr183a
|
G protein-coupled receptor 183a |
| chr12_+_34051848 | 2.03 |
ENSDART00000153276
|
cyth1b
|
cytohesin 1b |
| chr23_+_19198244 | 2.02 |
ENSDART00000047015
|
ccdc115
|
coiled-coil domain containing 115 |
| chr10_-_33588310 | 2.02 |
ENSDART00000108658
|
eva1c
|
eva-1 homolog C (C. elegans) |
| chr12_-_4540564 | 2.02 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr8_-_4596662 | 2.01 |
ENSDART00000138199
|
sept5a
|
septin 5a |
| chr18_-_6908434 | 2.00 |
ENSDART00000146536
|
si:dkey-266m15.6
|
si:dkey-266m15.6 |
| chr7_-_1348640 | 1.99 |
ENSDART00000130012
|
ripk3
|
receptor-interacting serine-threonine kinase 3 |
| chr8_-_49766205 | 1.98 |
ENSDART00000137941
ENSDART00000097919 ENSDART00000147309 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr4_+_75458537 | 1.97 |
ENSDART00000166278
|
si:ch211-266c8.1
|
si:ch211-266c8.1 |
| chr21_+_5993188 | 1.97 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr10_-_3138403 | 1.97 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr2_-_23768818 | 1.96 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr22_+_11857356 | 1.96 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr5_-_37117778 | 1.96 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr21_-_22317920 | 1.94 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr7_+_31132588 | 1.94 |
ENSDART00000173702
|
tjp1a
|
tight junction protein 1a |
| chr12_-_33746111 | 1.94 |
ENSDART00000127203
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr12_+_9499742 | 1.91 |
ENSDART00000044150
ENSDART00000136354 |
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr15_+_19884242 | 1.91 |
ENSDART00000154437
ENSDART00000054416 |
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr15_+_37936458 | 1.91 |
ENSDART00000154491
|
si:ch73-380l3.2
|
si:ch73-380l3.2 |
| chr1_+_27808916 | 1.91 |
ENSDART00000102335
|
pspc1
|
paraspeckle component 1 |
| chr18_+_15106518 | 1.90 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr10_-_33588106 | 1.90 |
ENSDART00000155634
|
eva1c
|
eva-1 homolog C (C. elegans) |
| chr5_-_57723929 | 1.89 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr3_+_32416948 | 1.89 |
ENSDART00000157324
ENSDART00000154267 ENSDART00000186094 ENSDART00000155860 ENSDART00000156986 |
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr18_-_40773413 | 1.88 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr15_+_5086338 | 1.88 |
ENSDART00000155402
|
mxf
|
myxovirus (influenza virus) resistance F |
| chr5_-_39805874 | 1.88 |
ENSDART00000176202
ENSDART00000191683 |
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr24_-_3426620 | 1.87 |
ENSDART00000184346
|
nck1b
|
NCK adaptor protein 1b |
| chr1_+_45085194 | 1.87 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr5_-_39805620 | 1.87 |
ENSDART00000137801
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr8_-_44224198 | 1.86 |
ENSDART00000132015
|
stx2b
|
syntaxin 2b |
| chr24_-_1341543 | 1.85 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr7_+_53154946 | 1.85 |
ENSDART00000185104
ENSDART00000191559 ENSDART00000160096 |
cdh29
|
cadherin 29 |
| chr7_+_8367929 | 1.85 |
ENSDART00000173303
|
zgc:172143
|
zgc:172143 |
| chr9_-_34509997 | 1.83 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr2_+_35595454 | 1.83 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr23_+_17839187 | 1.83 |
ENSDART00000104647
|
prim1
|
DNA primase subunit 1 |
| chr1_-_17587552 | 1.83 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr7_-_7984015 | 1.83 |
ENSDART00000163203
|
si:cabz01030277.1
|
si:cabz01030277.1 |
| chr15_-_17813680 | 1.82 |
ENSDART00000158556
|
CT573342.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of pou3f2a+pou3f2b+pou3f3a+pou3f3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.3 | GO:0002829 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.7 | 7.0 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 1.3 | 4.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.3 | 5.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 1.1 | 6.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 1.0 | 4.1 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 0.9 | 4.3 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.8 | 5.0 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.8 | 5.4 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.8 | 3.0 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.7 | 2.2 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.7 | 1.5 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.7 | 2.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.7 | 2.0 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.6 | 3.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.6 | 2.4 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.6 | 1.8 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.6 | 3.3 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.5 | 2.6 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.5 | 1.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.5 | 3.0 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.5 | 2.0 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.5 | 1.5 | GO:0032637 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.5 | 1.5 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.5 | 5.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.5 | 1.8 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 1.8 | GO:0014005 | microglia development(GO:0014005) |
| 0.4 | 1.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.4 | 1.3 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.4 | 2.6 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 1.7 | GO:0045217 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) cell-cell junction maintenance(GO:0045217) |
| 0.4 | 3.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 2.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.4 | 1.9 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.4 | 4.2 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 4.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.4 | 2.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.4 | 1.9 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.4 | 4.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 | 1.1 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.3 | 2.7 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.3 | 3.1 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.3 | 2.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 2.4 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.3 | 1.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.3 | 1.3 | GO:0050961 | thermoception(GO:0050955) detection of temperature stimulus involved in thermoception(GO:0050960) detection of temperature stimulus involved in sensory perception(GO:0050961) |
| 0.3 | 2.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 1.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 0.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.3 | 1.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.3 | 0.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 5.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.3 | 1.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.3 | 2.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 1.4 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.3 | 1.6 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.3 | 1.6 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.3 | 1.6 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 1.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.3 | 1.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.3 | 2.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 2.3 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.3 | 1.6 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.3 | 2.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 1.5 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 0.5 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.2 | 1.7 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 0.7 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.2 | 0.7 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 1.1 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.2 | 1.8 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 0.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 1.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 4.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 4.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 3.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 0.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 1.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.2 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.2 | 2.4 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 0.8 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.2 | 1.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 1.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 1.0 | GO:2000319 | regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.2 | 1.1 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.2 | 2.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 5.3 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.2 | 1.3 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.2 | 0.9 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 0.9 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 3.8 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.2 | 1.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.5 | GO:0036135 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.2 | 3.5 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.2 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 5.0 | GO:0007568 | aging(GO:0007568) |
| 0.2 | 0.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 3.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 1.8 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.2 | 1.3 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.2 | 1.0 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.2 | 0.8 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 1.6 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 5.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.2 | 1.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 0.6 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 1.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 0.5 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.2 | 0.9 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 4.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 1.0 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.4 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 1.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 2.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 2.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.8 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 3.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.9 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.5 | GO:0046823 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 1.0 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.1 | 1.7 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 2.4 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.1 | 1.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.6 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.1 | 3.8 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.1 | 0.9 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.6 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 1.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.5 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.7 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 2.0 | GO:0001843 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) tube closure(GO:0060606) |
| 0.1 | 0.3 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 2.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.6 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 3.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.8 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.4 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 1.2 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.9 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 0.8 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 3.1 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.1 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 3.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.2 | GO:0007260 | tyrosine phosphorylation of STAT protein(GO:0007260) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 2.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.1 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 1.8 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 5.3 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.1 | 1.2 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 3.0 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 1.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.4 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 1.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.5 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 2.9 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 1.4 | GO:0009749 | response to glucose(GO:0009749) |
| 0.1 | 1.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.4 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 2.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 5.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.5 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 5.7 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.1 | 1.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 1.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.7 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.1 | 1.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 1.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 2.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.8 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 1.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 3.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 0.6 | GO:0070836 | membrane raft assembly(GO:0001765) membrane raft organization(GO:0031579) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 7.2 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 1.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.8 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 1.7 | GO:0044236 | collagen metabolic process(GO:0032963) multicellular organism metabolic process(GO:0044236) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.1 | 0.2 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 2.2 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.4 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.1 | 1.1 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.0 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 1.0 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.6 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.3 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 1.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 1.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.2 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 6.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 1.4 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.6 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.0 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.0 | GO:0061386 | positive regulation of BMP signaling pathway(GO:0030513) closure of optic fissure(GO:0061386) |
| 0.0 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.0 | 0.7 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.9 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.0 | GO:1901215 | negative regulation of neuron death(GO:1901215) |
| 0.0 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 1.0 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 1.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 8.3 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.8 | GO:1904018 | positive regulation of angiogenesis(GO:0045766) positive regulation of vasculature development(GO:1904018) |
| 0.0 | 1.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.2 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 2.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 3.1 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.8 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 1.6 | GO:0036293 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.0 | 0.9 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.8 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 6.8 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.3 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0002793 | positive regulation of peptide secretion(GO:0002793) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.0 | 0.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.2 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 1.3 | GO:1902275 | regulation of chromatin organization(GO:1902275) |
| 0.0 | 0.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.9 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 1.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.5 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.6 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.1 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 1.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 7.8 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 1.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 2.5 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.1 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.4 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.9 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 17.9 | GO:0035556 | intracellular signal transduction(GO:0035556) |
| 0.0 | 1.7 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.9 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 1.0 | 14.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.7 | 5.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 2.8 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.7 | 3.5 | GO:0043034 | costamere(GO:0043034) |
| 0.6 | 1.8 | GO:1990077 | primosome complex(GO:1990077) |
| 0.4 | 1.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.4 | 4.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 4.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 1.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.4 | 1.4 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.4 | 5.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 0.7 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.3 | 0.9 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.3 | 1.7 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.3 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.7 | GO:0072380 | TRC complex(GO:0072380) |
| 0.2 | 0.9 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 1.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 0.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 1.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 1.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 4.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 1.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 1.0 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 1.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 1.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 3.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 2.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.2 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 0.5 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.1 | 1.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 3.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 7.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.7 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 3.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.7 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.8 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 4.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.5 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 0.3 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.1 | 0.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 8.5 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.7 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 5.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.8 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 1.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 5.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 2.0 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 4.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 3.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 9.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.3 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 1.5 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 3.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 2.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 12.9 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 12.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.9 | 9.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.3 | 3.9 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.9 | 2.7 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.6 | 3.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.6 | 5.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.6 | 2.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.6 | 2.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.6 | 2.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.5 | 2.2 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.5 | 5.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.5 | 2.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.5 | 1.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.5 | 1.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 1.3 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.4 | 4.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 4.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 5.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.4 | 4.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.4 | 4.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.4 | 1.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.3 | 2.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 1.0 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 2.4 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.3 | 1.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 1.3 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 0.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.3 | 1.2 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.3 | 1.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 2.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.3 | 1.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.3 | 1.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 1.1 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.3 | 0.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 2.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 2.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 4.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.3 | 1.3 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 7.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 1.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 2.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 3.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 3.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.2 | 1.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 1.1 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.2 | 2.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 0.6 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.2 | 2.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 0.9 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.2 | 3.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 1.6 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 2.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 1.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 0.9 | GO:0008832 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.2 | 1.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 0.8 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 1.0 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.8 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 3.9 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 2.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.2 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 2.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 8.1 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 1.0 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 1.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 5.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.4 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.1 | 1.0 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 2.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.0 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.8 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 1.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 3.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 3.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 2.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 0.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 4.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.7 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 2.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 2.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 4.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 4.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 1.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 7.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 2.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 2.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 4.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 3.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 7.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.5 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 1.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 6.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 3.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.5 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 1.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 13.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 4.4 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 18.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.3 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 2.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.6 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 3.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 1.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 2.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 2.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 1.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 3.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 5.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.7 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.1 | 4.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 6.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 3.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.5 | 4.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 5.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 4.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.3 | 5.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 4.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 3.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 1.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 1.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 1.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 1.7 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.2 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 3.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 5.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 2.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 3.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.0 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 3.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 4.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |