Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
Results for pitx2
Z-value: 1.19
Transcription factors associated with pitx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pitx2
|
ENSDARG00000036194 | paired-like homeodomain 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pitx2 | dr11_v1_chr14_+_36218072_36218072 | -0.11 | 2.8e-01 | Click! |
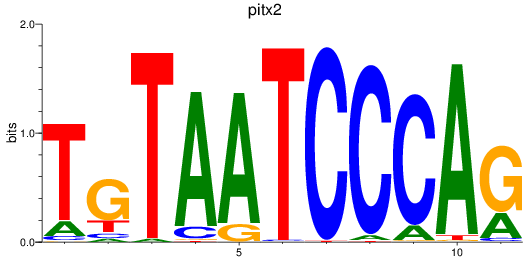
Activity profile of pitx2 motif
Sorted Z-values of pitx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_45510977 | 22.92 |
ENSDART00000090596
|
fgf12b
|
fibroblast growth factor 12b |
| chr12_+_47917971 | 12.06 |
ENSDART00000185933
|
tbata
|
thymus, brain and testes associated |
| chr13_+_24280380 | 9.33 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr5_+_24287927 | 7.26 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr9_-_34974143 | 6.91 |
ENSDART00000127295
|
zgc:172067
|
zgc:172067 |
| chr23_+_28731379 | 6.84 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr9_-_34974416 | 6.78 |
ENSDART00000077790
|
zgc:172067
|
zgc:172067 |
| chr19_+_12762887 | 6.18 |
ENSDART00000139909
|
mc5ra
|
melanocortin 5a receptor |
| chr21_+_22630627 | 6.15 |
ENSDART00000193092
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr25_+_6306885 | 6.02 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr14_-_2270973 | 5.81 |
ENSDART00000180729
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr21_+_22630297 | 5.53 |
ENSDART00000147175
|
si:dkeyp-69c1.7
|
si:dkeyp-69c1.7 |
| chr4_+_6643421 | 5.50 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr5_-_23277939 | 5.38 |
ENSDART00000003514
|
plp1b
|
proteolipid protein 1b |
| chr7_-_48173440 | 5.36 |
ENSDART00000124075
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr2_-_2020044 | 5.33 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr13_-_30028103 | 5.28 |
ENSDART00000183889
|
scdb
|
stearoyl-CoA desaturase b |
| chr5_-_20205075 | 5.17 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr25_-_13839743 | 4.97 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr24_+_34085940 | 4.93 |
ENSDART00000171189
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr5_-_72125551 | 4.72 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr9_-_43142636 | 4.67 |
ENSDART00000134349
ENSDART00000181835 |
ccdc141
|
coiled-coil domain containing 141 |
| chr19_-_9829965 | 4.66 |
ENSDART00000136842
ENSDART00000142766 |
cacng8a
|
calcium channel, voltage-dependent, gamma subunit 8a |
| chr7_-_57933736 | 4.62 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr11_-_4235811 | 4.60 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr23_-_24856025 | 4.59 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr1_+_7546259 | 4.40 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr23_+_40109353 | 4.24 |
ENSDART00000149249
|
ghrhrl
|
growth hormone releasing hormone receptor, like |
| chr21_+_10756154 | 4.20 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr1_+_33383644 | 4.17 |
ENSDART00000187194
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr9_+_307863 | 4.16 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr20_+_36233873 | 4.15 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr8_+_27807974 | 4.12 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr9_-_12034444 | 4.03 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr10_+_29963518 | 3.97 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr7_-_22132265 | 3.95 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr19_-_25271155 | 3.90 |
ENSDART00000104027
|
rims3
|
regulating synaptic membrane exocytosis 3 |
| chr8_+_554531 | 3.85 |
ENSDART00000193623
|
FO704758.2
|
|
| chr17_+_5915875 | 3.75 |
ENSDART00000184179
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr6_-_54126463 | 3.74 |
ENSDART00000161059
|
tusc2a
|
tumor suppressor candidate 2a |
| chr2_-_42871286 | 3.72 |
ENSDART00000087823
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr17_-_5583345 | 3.67 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr21_+_13366353 | 3.62 |
ENSDART00000151630
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr25_-_19090479 | 3.61 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr21_+_10866421 | 3.57 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr19_-_28283844 | 3.54 |
ENSDART00000151756
ENSDART00000079104 |
ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr2_-_6292510 | 3.52 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr4_+_8168514 | 3.41 |
ENSDART00000150830
|
ninj2
|
ninjurin 2 |
| chr18_+_26337869 | 3.40 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr9_-_32753535 | 3.38 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr16_-_43971258 | 3.36 |
ENSDART00000141941
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr13_-_24448278 | 3.35 |
ENSDART00000057584
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr11_+_6116503 | 3.35 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr23_+_40604951 | 3.34 |
ENSDART00000114959
|
cdh24a
|
cadherin 24, type 2a |
| chr24_+_24461341 | 3.28 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr10_-_43392267 | 3.23 |
ENSDART00000142872
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr10_+_15777064 | 3.22 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr1_-_1894722 | 3.22 |
ENSDART00000165669
|
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr14_-_2206476 | 3.20 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr16_-_13004166 | 3.18 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr6_+_45347219 | 3.16 |
ENSDART00000188240
|
LO017951.1
|
|
| chr10_+_15777258 | 3.13 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr24_-_27400630 | 3.13 |
ENSDART00000165760
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr9_-_29321625 | 3.11 |
ENSDART00000158689
ENSDART00000014047 ENSDART00000122602 |
pth2r
|
parathyroid hormone 2 receptor |
| chr14_+_44545092 | 3.10 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr13_+_16522608 | 3.06 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr15_-_12319065 | 3.06 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr13_+_52071971 | 3.04 |
ENSDART00000184775
|
CABZ01092745.1
|
|
| chr3_+_15296824 | 3.03 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr1_-_43987873 | 3.01 |
ENSDART00000108821
|
CR385050.1
|
|
| chr10_-_7857494 | 3.00 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr23_+_8989168 | 3.00 |
ENSDART00000034380
|
xkr7
|
XK, Kell blood group complex subunit-related family, member 7 |
| chr21_-_32467099 | 2.98 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr10_+_21511495 | 2.92 |
ENSDART00000178395
|
BX957322.3
|
|
| chr23_+_22267374 | 2.89 |
ENSDART00000079035
|
rap1gap
|
RAP1 GTPase activating protein |
| chr25_+_35019693 | 2.89 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr10_+_26926654 | 2.81 |
ENSDART00000078980
ENSDART00000100289 |
rab1bb
|
RAB1B, member RAS oncogene family b |
| chr18_+_48608366 | 2.76 |
ENSDART00000151229
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr12_+_48216662 | 2.76 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr6_-_36182115 | 2.68 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr21_+_41743493 | 2.64 |
ENSDART00000192669
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr18_-_21806613 | 2.61 |
ENSDART00000145721
|
nrn1la
|
neuritin 1-like a |
| chr5_+_24282570 | 2.58 |
ENSDART00000041905
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr9_+_33216945 | 2.56 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr6_-_6248893 | 2.53 |
ENSDART00000124662
|
rtn4a
|
reticulon 4a |
| chr7_+_24115082 | 2.48 |
ENSDART00000182718
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr23_+_33934228 | 2.47 |
ENSDART00000134237
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr5_-_38094130 | 2.46 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr20_-_18731268 | 2.46 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr23_-_24825863 | 2.45 |
ENSDART00000112493
|
syt6a
|
synaptotagmin VIa |
| chr17_-_43552894 | 2.45 |
ENSDART00000181226
ENSDART00000188125 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr25_-_7999756 | 2.44 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr13_-_30149973 | 2.41 |
ENSDART00000041515
|
sar1ab
|
secretion associated, Ras related GTPase 1Ab |
| chr5_-_67145505 | 2.40 |
ENSDART00000011295
|
rom1a
|
retinal outer segment membrane protein 1a |
| chr5_+_37978501 | 2.39 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr1_+_38758261 | 2.38 |
ENSDART00000182756
|
wdr17
|
WD repeat domain 17 |
| chr17_+_33495194 | 2.37 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr9_-_23747264 | 2.34 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr5_+_52067723 | 2.33 |
ENSDART00000166902
|
setbp1
|
SET binding protein 1 |
| chr7_+_15872357 | 2.33 |
ENSDART00000165757
|
pax6b
|
paired box 6b |
| chr20_+_41640687 | 2.32 |
ENSDART00000138686
|
fam184a
|
family with sequence similarity 184, member A |
| chr13_-_30027730 | 2.31 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr16_-_26820634 | 2.27 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr2_-_31767827 | 2.25 |
ENSDART00000114928
|
and2
|
actinodin2 |
| chr5_-_29488245 | 2.25 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr1_+_38758445 | 2.24 |
ENSDART00000136300
|
wdr17
|
WD repeat domain 17 |
| chr11_-_44409856 | 2.22 |
ENSDART00000162886
|
il1rapl1b
|
interleukin 1 receptor accessory protein-like 1b |
| chr1_+_11881559 | 2.20 |
ENSDART00000166981
|
snx8b
|
sorting nexin 8b |
| chr9_-_42989297 | 2.20 |
ENSDART00000126871
|
ttn.2
|
titin, tandem duplicate 2 |
| chr14_-_2264494 | 2.19 |
ENSDART00000191149
|
pcdh2ab9
|
protocadherin 2 alpha b 9 |
| chr20_-_29482492 | 2.18 |
ENSDART00000178308
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr19_+_37701450 | 2.18 |
ENSDART00000087694
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr19_-_40776267 | 2.17 |
ENSDART00000189038
|
calcr
|
calcitonin receptor |
| chr18_+_9323211 | 2.17 |
ENSDART00000166114
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr25_+_25124684 | 2.15 |
ENSDART00000167542
|
ldha
|
lactate dehydrogenase A4 |
| chr17_-_47142249 | 2.13 |
ENSDART00000184705
|
CU915775.1
|
|
| chr20_-_30369598 | 2.12 |
ENSDART00000144549
|
allc
|
allantoicase |
| chr7_+_30970045 | 2.11 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr7_-_65191937 | 2.10 |
ENSDART00000173234
|
pkd1l2a
|
polycystic kidney disease 1 like 2a |
| chr22_-_25469751 | 2.09 |
ENSDART00000171670
|
CR769772.4
|
|
| chr8_+_50983551 | 2.06 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr21_+_20715020 | 2.06 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr6_-_9581949 | 2.06 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr11_-_28148033 | 2.05 |
ENSDART00000177182
|
lactbl1b
|
lactamase, beta-like 1b |
| chr21_+_21738150 | 2.05 |
ENSDART00000150974
|
or124-4
|
odorant receptor, family E, subfamily 124, member 4 |
| chr8_+_7737062 | 2.05 |
ENSDART00000166712
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr24_+_2470061 | 2.02 |
ENSDART00000140383
ENSDART00000191261 |
F13A1 (1 of many)
|
coagulation factor XIII A chain |
| chr25_+_35020529 | 2.01 |
ENSDART00000158016
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr17_-_17759138 | 2.01 |
ENSDART00000157128
ENSDART00000123845 |
adck1
|
aarF domain containing kinase 1 |
| chr23_+_9867483 | 2.00 |
ENSDART00000023099
|
slc16a7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2) |
| chr21_-_21790372 | 1.99 |
ENSDART00000151094
|
xrra1
|
X-ray radiation resistance associated 1 |
| chr10_-_25204034 | 1.98 |
ENSDART00000161100
|
FAT3 (1 of many)
|
si:ch211-214k5.6 |
| chr19_+_19747430 | 1.98 |
ENSDART00000166129
|
hoxa9a
|
homeobox A9a |
| chr19_+_42898239 | 1.96 |
ENSDART00000051724
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr10_-_25823258 | 1.96 |
ENSDART00000064327
|
ftr54
|
finTRIM family, member 54 |
| chr14_+_743346 | 1.94 |
ENSDART00000110511
|
klb
|
klotho beta |
| chr9_-_1986014 | 1.93 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr8_+_35356944 | 1.93 |
ENSDART00000143243
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr20_+_18225329 | 1.93 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr11_+_6116096 | 1.93 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr24_-_10006158 | 1.92 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr7_+_29167744 | 1.91 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr1_-_57129179 | 1.90 |
ENSDART00000157226
ENSDART00000152469 |
si:ch73-94k4.2
|
si:ch73-94k4.2 |
| chr6_+_37894914 | 1.87 |
ENSDART00000148817
|
oca2
|
oculocutaneous albinism II |
| chr11_-_16093018 | 1.87 |
ENSDART00000139309
ENSDART00000139819 |
SPATA1
|
si:dkey-205k8.5 |
| chr11_-_1933793 | 1.86 |
ENSDART00000186850
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr22_+_696931 | 1.84 |
ENSDART00000149712
ENSDART00000009756 |
gpr37l1a
|
G protein-coupled receptor 37 like 1a |
| chr10_+_8534750 | 1.84 |
ENSDART00000183960
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr15_-_31390985 | 1.84 |
ENSDART00000060117
|
or111-6
|
odorant receptor, family D, subfamily 111, member 6 |
| chr4_+_5848229 | 1.83 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr23_+_43849190 | 1.83 |
ENSDART00000017375
|
cnga1
|
cyclic nucleotide gated channel alpha 1 |
| chr9_-_53537989 | 1.83 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
| chr12_+_36416173 | 1.81 |
ENSDART00000190278
|
unk
|
unkempt family zinc finger |
| chr2_+_6253246 | 1.80 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr20_-_20533865 | 1.79 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr11_-_29563437 | 1.78 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr13_+_9896845 | 1.76 |
ENSDART00000169076
|
si:ch211-117n7.8
|
si:ch211-117n7.8 |
| chr17_-_50331351 | 1.76 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr6_-_39903393 | 1.72 |
ENSDART00000085945
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr4_-_40883162 | 1.70 |
ENSDART00000151978
|
znf1102
|
zinc finger protein 1102 |
| chr18_-_6534516 | 1.68 |
ENSDART00000009217
|
ddx11
|
DEAD/H (Asp-Glu-Ala-Asp/His) box helicase 11 |
| chr7_+_53413445 | 1.67 |
ENSDART00000190480
|
znf609b
|
zinc finger protein 609b |
| chr2_+_38731696 | 1.66 |
ENSDART00000181733
|
FQ377660.1
|
|
| chr11_-_3574311 | 1.66 |
ENSDART00000167064
|
si:dkey-33m11.7
|
si:dkey-33m11.7 |
| chr18_+_21061216 | 1.65 |
ENSDART00000141739
|
fam169b
|
family with sequence similarity 169, member B |
| chr12_+_28995942 | 1.65 |
ENSDART00000076334
|
valopb
|
vertebrate ancient long opsin b |
| chr7_-_16204885 | 1.65 |
ENSDART00000171008
|
btr05
|
bloodthirsty-related gene family, member 5 |
| chr12_+_316238 | 1.64 |
ENSDART00000187492
|
rcvrnb
|
recoverin b |
| chr8_-_22157301 | 1.61 |
ENSDART00000158383
|
nphp4
|
nephronophthisis 4 |
| chr1_-_26293203 | 1.59 |
ENSDART00000180140
|
cxxc4
|
CXXC finger 4 |
| chr4_+_38002643 | 1.58 |
ENSDART00000160860
|
znf1051
|
zinc finger protein 1051 |
| chr6_+_21740672 | 1.58 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr22_-_25502977 | 1.57 |
ENSDART00000181749
|
CR769772.4
|
|
| chr5_+_64840656 | 1.56 |
ENSDART00000073953
|
lrrc8ab
|
leucine rich repeat containing 8 VRAC subunit Ab |
| chr17_-_53328043 | 1.56 |
ENSDART00000171082
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr23_-_27505825 | 1.54 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr2_+_24931677 | 1.54 |
ENSDART00000021528
|
agtr1a
|
angiotensin II receptor, type 1a |
| chr8_-_36125849 | 1.54 |
ENSDART00000159581
|
CT583723.1
|
|
| chr20_+_25581627 | 1.52 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr17_-_50040927 | 1.51 |
ENSDART00000184304
|
FO834825.1
|
|
| chr2_-_11258547 | 1.50 |
ENSDART00000165803
ENSDART00000193817 |
slc44a5a
|
solute carrier family 44, member 5a |
| chr25_-_35047838 | 1.50 |
ENSDART00000155060
|
CU302436.3
|
|
| chr2_-_2096055 | 1.49 |
ENSDART00000126566
|
slc22a23
|
solute carrier family 22, member 23 |
| chr8_+_41533268 | 1.49 |
ENSDART00000142377
|
si:ch211-158d24.2
|
si:ch211-158d24.2 |
| chr17_-_16342388 | 1.48 |
ENSDART00000017930
|
kcnk13a
|
potassium channel, subfamily K, member 13a |
| chr7_+_24114694 | 1.47 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr18_-_39702327 | 1.46 |
ENSDART00000149158
|
dmxl2
|
Dmx-like 2 |
| chr11_-_43002262 | 1.46 |
ENSDART00000172477
ENSDART00000181513 |
CABZ01069998.1
|
|
| chr16_-_45910050 | 1.46 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr10_+_36150911 | 1.45 |
ENSDART00000076804
|
waif2
|
Wnt-activated inhibitory factor 2 |
| chr1_-_10577945 | 1.45 |
ENSDART00000179237
ENSDART00000040502 ENSDART00000186876 |
trpc5a
|
transient receptor potential cation channel, subfamily C, member 5a |
| chr6_+_18520859 | 1.44 |
ENSDART00000158263
|
si:dkey-10p5.10
|
si:dkey-10p5.10 |
| chr23_+_3596401 | 1.44 |
ENSDART00000185908
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr14_+_41518257 | 1.42 |
ENSDART00000050037
|
chrnb3b
|
cholinergic receptor, nicotinic, beta 3b (neuronal) |
| chr21_+_13127742 | 1.40 |
ENSDART00000179221
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr15_-_37929135 | 1.40 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr14_-_41535822 | 1.39 |
ENSDART00000149407
|
itga6l
|
integrin, alpha 6, like |
| chr7_+_42208859 | 1.38 |
ENSDART00000148643
|
phkb
|
phosphorylase kinase, beta |
| chr8_+_694218 | 1.37 |
ENSDART00000147753
|
rnf165b
|
ring finger protein 165b |
| chr21_-_33032715 | 1.35 |
ENSDART00000065349
ENSDART00000191502 ENSDART00000149386 |
slc34a1b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of pitx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 1.6 | 22.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.4 | 4.2 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 1.3 | 5.2 | GO:0046144 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 1.2 | 6.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 1.0 | 2.9 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.9 | 4.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.8 | 3.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.8 | 4.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.7 | 3.7 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.7 | 2.1 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.7 | 13.7 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.6 | 2.4 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.6 | 1.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.6 | 2.3 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.5 | 4.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.5 | 5.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.5 | 2.5 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.5 | 1.5 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.5 | 2.4 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.5 | 7.8 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.4 | 2.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.4 | 2.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.4 | 2.8 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.4 | 1.6 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.4 | 3.1 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.4 | 3.0 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.4 | 4.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.4 | 2.2 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.4 | 2.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.4 | 3.9 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.3 | 1.0 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 3.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 5.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 1.9 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 1.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 3.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.3 | 3.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.3 | 5.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.3 | 2.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.3 | 8.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.3 | 5.4 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.3 | 3.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.3 | 2.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.0 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 | 4.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 8.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 3.6 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 1.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 2.6 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.2 | 1.9 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 2.2 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 2.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 1.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.2 | 2.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 2.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 3.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.2 | 0.3 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.2 | 22.5 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.2 | 4.7 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 0.7 | GO:0061440 | renal system vasculature development(GO:0061437) kidney vasculature development(GO:0061440) glomerulus vasculature development(GO:0072012) |
| 0.1 | 1.8 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 2.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 8.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.5 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 9.3 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 3.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 0.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.7 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.6 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.4 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 4.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.9 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 2.0 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 3.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.9 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.5 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 1.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 3.3 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.1 | 0.6 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 5.0 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.1 | 1.0 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 1.5 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.1 | 0.6 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 2.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 14.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 3.1 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.1 | 0.7 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.7 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.1 | 4.6 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 1.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.3 | GO:0033135 | regulation of peptidyl-serine phosphorylation(GO:0033135) positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 3.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.9 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 1.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 1.5 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.5 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 2.3 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 1.0 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.9 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 2.4 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 9.7 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 9.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.1 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.9 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.8 | GO:0099590 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 1.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 4.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 2.4 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 2.0 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 1.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 2.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 7.1 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.4 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.9 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.7 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.7 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.3 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 5.7 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 2.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.0 | 4.3 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) trans-synaptic signaling(GO:0099537) |
| 0.0 | 2.1 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 7.4 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.9 | GO:0021782 | glial cell development(GO:0021782) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.0 | 4.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.5 | 8.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.5 | 8.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.4 | 1.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 1.6 | GO:1990923 | PET complex(GO:1990923) |
| 0.3 | 5.9 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 2.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 7.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 6.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.9 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 1.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.0 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 2.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 7.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 2.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 8.8 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 1.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 2.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.4 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.9 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 9.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 4.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.9 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 2.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 8.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 5.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 2.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 4.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 5.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 2.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 8.8 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 1.3 | 5.2 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 1.0 | 8.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 1.0 | 26.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.7 | 2.2 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.7 | 2.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.7 | 4.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.7 | 6.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.6 | 1.8 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.6 | 2.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 2.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.4 | 1.9 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.4 | 1.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.4 | 3.0 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.3 | 2.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 2.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 3.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.3 | 6.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 1.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.0 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.2 | 4.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 2.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 2.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 1.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 2.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 2.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 11.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 8.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 16.2 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 3.1 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 14.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 1.2 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.2 | 4.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.8 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 1.9 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 3.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.4 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.1 | 1.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 3.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 2.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 2.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 7.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 3.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.9 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 3.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.2 | GO:0016849 | phosphorus-oxygen lyase activity(GO:0016849) |
| 0.0 | 7.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 5.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 4.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 2.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.9 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 2.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 2.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 4.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 4.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 3.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 7.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 10.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.7 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 1.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.7 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 3.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.3 | 2.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 2.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 3.7 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.2 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 1.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 4.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 2.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 3.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 6.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 5.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.9 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.1 | 0.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |