Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
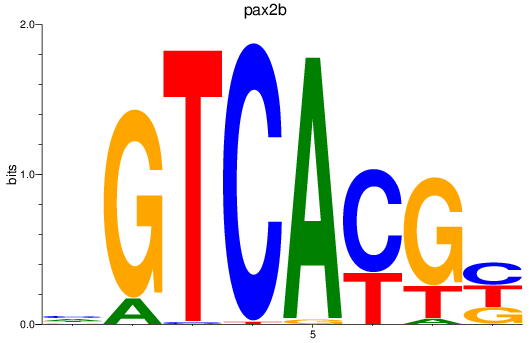
Results for pax2b
Z-value: 1.56
Transcription factors associated with pax2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pax2b
|
ENSDARG00000032578 | paired box 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pax2b | dr11_v1_chr12_-_45876387_45876387 | 0.39 | 1.1e-04 | Click! |
Activity profile of pax2b motif
Sorted Z-values of pax2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_10720294 | 10.70 |
ENSDART00000139387
|
atp1b1b
|
ATPase Na+/K+ transporting subunit beta 1b |
| chr16_-_45069882 | 8.56 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr11_+_25112269 | 7.73 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr22_-_11136625 | 7.30 |
ENSDART00000016873
ENSDART00000125561 |
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr2_-_38000276 | 7.08 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr7_+_38770167 | 6.91 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr24_-_7632187 | 6.73 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr7_-_42206720 | 6.50 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr23_-_4915118 | 6.37 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr24_-_17023392 | 6.36 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr24_-_6158933 | 6.36 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr15_+_28989573 | 6.28 |
ENSDART00000076648
|
clip3
|
CAP-GLY domain containing linker protein 3 |
| chr18_-_7137153 | 6.17 |
ENSDART00000019571
|
cd9a
|
CD9 molecule a |
| chr11_-_10798021 | 5.33 |
ENSDART00000167112
ENSDART00000179725 ENSDART00000091923 ENSDART00000185825 |
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr7_-_30082931 | 5.14 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr16_+_10777116 | 5.05 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr21_-_39628771 | 4.99 |
ENSDART00000183995
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr3_-_32362872 | 4.94 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr8_-_25002182 | 4.94 |
ENSDART00000078792
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr24_+_31334209 | 4.69 |
ENSDART00000168837
ENSDART00000172473 |
fam168b
|
family with sequence similarity 168, member B |
| chr15_-_163586 | 4.62 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr4_-_5302866 | 4.58 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr12_+_16953415 | 4.52 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr21_+_13861589 | 4.47 |
ENSDART00000015629
ENSDART00000171306 |
stxbp1a
|
syntaxin binding protein 1a |
| chr20_+_20637866 | 4.46 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr4_+_9669717 | 4.42 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr2_+_42871831 | 4.41 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr25_-_21156678 | 4.40 |
ENSDART00000156257
|
wnk1a
|
WNK lysine deficient protein kinase 1a |
| chr2_-_33687214 | 4.36 |
ENSDART00000147439
|
atp6v0b
|
ATPase H+ transporting V0 subunit b |
| chr7_-_44605050 | 4.36 |
ENSDART00000148471
ENSDART00000149072 |
tk2
|
thymidine kinase 2, mitochondrial |
| chr10_+_15777258 | 4.31 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr12_-_33972798 | 4.30 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr8_+_25761654 | 4.27 |
ENSDART00000137899
ENSDART00000062403 |
tmem9
|
transmembrane protein 9 |
| chr2_-_39759059 | 4.16 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr14_-_24410673 | 4.16 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr8_+_26292560 | 4.13 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr7_+_44715224 | 3.97 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr18_+_43183749 | 3.92 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr2_-_31302615 | 3.92 |
ENSDART00000034784
ENSDART00000060812 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr14_+_22172047 | 3.91 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr8_+_16758304 | 3.90 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr7_-_32895668 | 3.90 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr19_+_33093395 | 3.89 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr24_+_39158283 | 3.82 |
ENSDART00000053139
|
atp6v0cb
|
ATPase H+ transporting V0 subunit cb |
| chr19_-_42416696 | 3.78 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr20_+_29743904 | 3.75 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr15_-_20956384 | 3.75 |
ENSDART00000135770
|
tbcela
|
tubulin folding cofactor E-like a |
| chr3_+_40576447 | 3.74 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr21_+_22400951 | 3.69 |
ENSDART00000134592
|
lmbrd2b
|
LMBR1 domain containing 2b |
| chr11_+_37275448 | 3.68 |
ENSDART00000161423
|
creld1a
|
cysteine-rich with EGF-like domains 1a |
| chr20_+_27393668 | 3.65 |
ENSDART00000005473
|
tmem179
|
transmembrane protein 179 |
| chr16_+_25761101 | 3.62 |
ENSDART00000110619
|
cblc
|
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr16_+_23960744 | 3.60 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr13_+_421231 | 3.56 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr3_-_15264698 | 3.56 |
ENSDART00000111948
ENSDART00000142594 |
sez6l2
|
seizure related 6 homolog (mouse)-like 2 |
| chr25_-_13871118 | 3.55 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr1_+_45707219 | 3.54 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr16_+_10841163 | 3.53 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr25_+_13791627 | 3.51 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr6_+_48041759 | 3.50 |
ENSDART00000140086
|
si:dkey-92f12.2
|
si:dkey-92f12.2 |
| chr24_-_33756003 | 3.49 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr10_+_16501699 | 3.48 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr20_-_20610812 | 3.45 |
ENSDART00000181870
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr2_+_30786773 | 3.40 |
ENSDART00000019029
ENSDART00000145681 |
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr1_-_10473630 | 3.38 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr18_+_40381102 | 3.37 |
ENSDART00000136588
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr10_+_23060391 | 3.36 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr9_+_17429170 | 3.34 |
ENSDART00000006256
|
zgc:101559
|
zgc:101559 |
| chr2_-_24996441 | 3.34 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr3_+_27713610 | 3.32 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr4_+_73973242 | 3.32 |
ENSDART00000182529
|
LO018181.1
|
|
| chr16_+_23960933 | 3.31 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr15_+_16387088 | 3.31 |
ENSDART00000101789
|
flot2b
|
flotillin 2b |
| chr23_+_24973773 | 3.26 |
ENSDART00000047020
|
casp9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr13_-_23007813 | 3.26 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr13_+_11439486 | 3.25 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr10_+_20108557 | 3.25 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| chr19_-_26823647 | 3.24 |
ENSDART00000002464
|
neu1
|
neuraminidase 1 |
| chr5_+_33301005 | 3.23 |
ENSDART00000006021
|
usp20
|
ubiquitin specific peptidase 20 |
| chr22_-_21176269 | 3.23 |
ENSDART00000112839
|
rex1bd
|
required for excision 1-B domain containing |
| chr2_+_30787128 | 3.21 |
ENSDART00000189233
|
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr24_+_12989727 | 3.20 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr18_+_26337869 | 3.20 |
ENSDART00000109257
|
RASGRF1
|
si:ch211-234p18.3 |
| chr17_+_5895500 | 3.20 |
ENSDART00000019905
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr4_-_5247335 | 3.19 |
ENSDART00000050221
|
atp6v1e1b
|
ATPase H+ transporting V1 subunit E1b |
| chr23_+_44741500 | 3.18 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr2_-_30460293 | 3.17 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr15_+_32798333 | 3.17 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr1_-_46924801 | 3.17 |
ENSDART00000142560
|
pdxkb
|
pyridoxal (pyridoxine, vitamin B6) kinase b |
| chr8_+_31248917 | 3.16 |
ENSDART00000112170
|
unm_hu7912
|
un-named hu7912 |
| chr24_-_4148914 | 3.15 |
ENSDART00000033494
ENSDART00000147849 |
klf6a
|
Kruppel-like factor 6a |
| chr14_+_14847304 | 3.13 |
ENSDART00000169932
|
ccdc142
|
coiled-coil domain containing 142 |
| chr5_+_57320113 | 3.12 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr18_+_17428506 | 3.11 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr11_+_5817202 | 3.07 |
ENSDART00000126084
|
ctdspl3
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 3 |
| chr23_-_29505463 | 3.06 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr15_-_19250543 | 3.05 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr19_+_551963 | 3.03 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr11_+_5499661 | 3.03 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr1_+_45671687 | 3.01 |
ENSDART00000146101
|
mcoln1a
|
mucolipin 1a |
| chr17_+_21964472 | 3.00 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr3_-_37785873 | 2.99 |
ENSDART00000011691
|
baxa
|
BCL2 associated X, apoptosis regulator a |
| chr24_-_10828560 | 2.95 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr4_+_16885854 | 2.94 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr19_-_20114149 | 2.93 |
ENSDART00000052620
|
npy
|
neuropeptide Y |
| chr21_+_22878991 | 2.88 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr19_-_12193622 | 2.88 |
ENSDART00000041960
|
ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr1_-_31505144 | 2.87 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr3_-_21062706 | 2.87 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr19_+_33093577 | 2.86 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr10_-_7702029 | 2.85 |
ENSDART00000018017
|
gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr9_-_1703761 | 2.85 |
ENSDART00000144822
ENSDART00000137210 ENSDART00000135273 |
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr23_+_37086159 | 2.85 |
ENSDART00000074407
|
cptp
|
ceramide-1-phosphate transfer protein |
| chr16_+_14216581 | 2.84 |
ENSDART00000113093
|
gba
|
glucosidase, beta, acid |
| chr13_+_35925490 | 2.82 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr23_-_16692312 | 2.81 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr10_+_25219728 | 2.81 |
ENSDART00000193829
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr7_-_28696556 | 2.81 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr5_+_11812089 | 2.80 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr11_-_22372072 | 2.80 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr21_+_22878834 | 2.79 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr18_-_22094102 | 2.79 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr22_-_26852516 | 2.77 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr9_+_34641237 | 2.73 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr19_-_17774875 | 2.73 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr16_-_16237844 | 2.72 |
ENSDART00000168747
ENSDART00000111912 |
rbm12b
|
RNA binding motif protein 12B |
| chr12_+_24342303 | 2.72 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr7_-_41013575 | 2.71 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr18_+_6857071 | 2.69 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr11_-_1291012 | 2.68 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr10_-_7974155 | 2.67 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr3_-_21106093 | 2.67 |
ENSDART00000156566
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr21_-_42100471 | 2.66 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr2_+_301898 | 2.66 |
ENSDART00000157246
|
znf1008
|
zinc finger protein 1008 |
| chr22_+_17399124 | 2.66 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr9_+_21819082 | 2.66 |
ENSDART00000136902
ENSDART00000101991 |
txndc9
|
thioredoxin domain containing 9 |
| chr25_+_1591964 | 2.65 |
ENSDART00000093277
|
ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr9_+_28103735 | 2.64 |
ENSDART00000007789
|
idh1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr7_+_57795974 | 2.64 |
ENSDART00000148369
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr1_+_7956030 | 2.64 |
ENSDART00000159655
|
CR855320.1
|
|
| chr2_+_33189582 | 2.63 |
ENSDART00000145588
ENSDART00000136330 ENSDART00000139295 ENSDART00000086340 |
rnf220a
|
ring finger protein 220a |
| chr23_-_12345764 | 2.63 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr13_+_2357637 | 2.63 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr2_-_54387550 | 2.62 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr6_+_27667359 | 2.61 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr8_+_26083808 | 2.60 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr1_+_47091468 | 2.60 |
ENSDART00000036783
|
cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr15_+_28685892 | 2.58 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr16_+_31853919 | 2.58 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr19_+_8985230 | 2.57 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr5_-_22130937 | 2.56 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr23_+_33718602 | 2.55 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr14_+_8947282 | 2.55 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr17_-_50010121 | 2.53 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr8_-_30944465 | 2.52 |
ENSDART00000128792
ENSDART00000191717 ENSDART00000049944 |
smarcb1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1a |
| chr19_-_5103313 | 2.52 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr16_-_2558653 | 2.51 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr8_-_44904723 | 2.51 |
ENSDART00000040804
|
praf2
|
PRA1 domain family, member 2 |
| chr13_-_45022527 | 2.51 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr2_-_31614277 | 2.51 |
ENSDART00000137054
ENSDART00000131507 ENSDART00000137674 ENSDART00000147041 |
si:ch211-106h4.5
|
si:ch211-106h4.5 |
| chr3_+_32832538 | 2.50 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr7_+_42206847 | 2.49 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr19_-_5103141 | 2.49 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr9_-_18743012 | 2.48 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr21_+_21743599 | 2.47 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr2_+_41526904 | 2.47 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr14_+_597532 | 2.46 |
ENSDART00000159805
|
LO018568.2
|
|
| chr20_-_26060154 | 2.44 |
ENSDART00000151950
|
serac1
|
serine active site containing 1 |
| chr4_-_16836006 | 2.42 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr17_-_7861219 | 2.42 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr7_+_26224211 | 2.40 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr7_-_28647959 | 2.38 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr13_-_33022372 | 2.38 |
ENSDART00000147165
|
rbm25a
|
RNA binding motif protein 25a |
| chr5_-_41103583 | 2.38 |
ENSDART00000051070
ENSDART00000074781 |
golph3
|
golgi phosphoprotein 3 |
| chr2_+_42177113 | 2.36 |
ENSDART00000056441
|
tmeff1a
|
transmembrane protein with EGF-like and two follistatin-like domains 1a |
| chr1_-_45614318 | 2.35 |
ENSDART00000149725
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_-_26191960 | 2.34 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr5_-_19006290 | 2.33 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr25_-_19443421 | 2.33 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr18_+_26086803 | 2.32 |
ENSDART00000187911
|
znf710a
|
zinc finger protein 710a |
| chr13_+_25505580 | 2.32 |
ENSDART00000140634
|
inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr2_+_20430366 | 2.31 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr2_+_12255568 | 2.30 |
ENSDART00000184164
ENSDART00000013454 |
prtfdc1
|
phosphoribosyl transferase domain containing 1 |
| chr23_-_35483163 | 2.30 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr7_-_41812015 | 2.30 |
ENSDART00000174058
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr23_-_36724575 | 2.29 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr8_+_24854600 | 2.28 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr3_-_50277959 | 2.28 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr22_+_16759010 | 2.27 |
ENSDART00000079638
ENSDART00000113099 |
tm2d1
|
TM2 domain containing 1 |
| chr1_+_6135176 | 2.27 |
ENSDART00000092324
ENSDART00000179970 |
abcb6a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6a |
| chr13_-_21701323 | 2.27 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr14_+_38786298 | 2.27 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr2_-_51096647 | 2.26 |
ENSDART00000167172
|
THEM6
|
si:ch73-52e5.2 |
| chr8_+_50983551 | 2.26 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr15_+_22311803 | 2.26 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr15_-_18138607 | 2.25 |
ENSDART00000176690
|
CR385077.1
|
|
| chr8_-_31716302 | 2.25 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr18_+_30028637 | 2.24 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr21_+_7131970 | 2.21 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr21_+_13205859 | 2.21 |
ENSDART00000102253
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pax2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 2.1 | 6.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.4 | 4.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 1.3 | 3.9 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 1.3 | 3.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 1.3 | 5.0 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.2 | 4.9 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 1.1 | 3.4 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 1.1 | 3.2 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 1.0 | 3.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 1.0 | 18.9 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.9 | 2.8 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.9 | 2.8 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.9 | 2.7 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.9 | 2.6 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.9 | 5.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.8 | 3.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.8 | 3.4 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.8 | 2.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.8 | 4.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.8 | 2.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.7 | 10.5 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.7 | 3.7 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.7 | 5.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.7 | 4.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.7 | 6.6 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.7 | 2.9 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.7 | 2.1 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.7 | 2.8 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.7 | 4.8 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.7 | 2.0 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.7 | 2.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.6 | 1.9 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.6 | 2.5 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.6 | 1.7 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.6 | 1.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.5 | 2.7 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.5 | 3.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.5 | 2.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 1.6 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.5 | 1.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.5 | 2.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.5 | 1.6 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.5 | 3.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.5 | 2.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.5 | 2.0 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.5 | 2.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 1.9 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.5 | 1.9 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.5 | 1.4 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.5 | 1.9 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.5 | 1.9 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.5 | 1.9 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.5 | 2.8 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.5 | 2.3 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.5 | 2.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.4 | 1.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 2.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.4 | 1.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.4 | 2.2 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.4 | 5.7 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 | 3.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.4 | 2.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.4 | 4.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.4 | 2.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.4 | 1.6 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.4 | 1.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 3.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.4 | 3.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 1.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.4 | 1.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 1.8 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.4 | 1.5 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.4 | 1.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.4 | 2.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 1.8 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.3 | 1.0 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.3 | 2.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.3 | 5.4 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 | 1.0 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.3 | 3.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.3 | 1.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 4.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 1.3 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.3 | 2.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 10.4 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.3 | 8.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 0.9 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.3 | 1.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 0.9 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.3 | 1.5 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.3 | 1.7 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.3 | 0.9 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.3 | 2.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 | 1.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.3 | 1.4 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.3 | 1.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.3 | 3.3 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.3 | 1.6 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.3 | 13.0 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.3 | 3.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 0.8 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.3 | 1.9 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.3 | 1.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.3 | 1.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.3 | 1.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 2.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.3 | 1.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.3 | 3.8 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.3 | 1.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 0.7 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 0.5 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.0 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.2 | 5.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 1.9 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 2.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.2 | 1.4 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.9 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 1.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 0.9 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 3.3 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.2 | 2.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.3 | GO:0070293 | renal absorption(GO:0070293) |
| 0.2 | 1.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 0.6 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.2 | 0.9 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.2 | 1.3 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.2 | 0.8 | GO:0071632 | optomotor response(GO:0071632) |
| 0.2 | 1.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 0.6 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.2 | 1.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.2 | 1.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 1.9 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.2 | 3.9 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.2 | 1.3 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 0.7 | GO:0021855 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamus cell migration(GO:0021855) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.2 | 0.9 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 0.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 2.0 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.2 | 1.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.2 | 5.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.9 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 0.9 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 1.0 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.2 | 3.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 3.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 0.5 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 2.0 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.2 | 0.7 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 0.5 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 1.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 0.5 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.2 | 3.9 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.2 | 0.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 2.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.5 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.2 | 1.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.2 | 1.7 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 4.0 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.2 | 1.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 0.5 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.2 | 1.2 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.2 | 1.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.2 | 0.9 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.7 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.9 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 3.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 2.3 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.9 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 2.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 8.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 2.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 4.8 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 4.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.1 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 1.8 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 2.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 3.6 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 3.4 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 4.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 4.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 3.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.6 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 2.8 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.1 | 1.8 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 2.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 6.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 3.5 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.1 | 1.0 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 2.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.3 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.1 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.5 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.1 | 1.1 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.7 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 2.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.6 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 2.0 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.6 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.7 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 3.6 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.1 | 0.6 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 2.1 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 1.8 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.1 | 2.2 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 4.4 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 0.3 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.6 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.1 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 1.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 1.0 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 4.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 2.2 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 2.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.5 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 1.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.9 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 6.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 1.4 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.1 | 0.4 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 2.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 4.0 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 3.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 6.5 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 2.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.7 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 0.8 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.8 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 1.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 2.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.3 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.1 | 2.0 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 1.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 3.7 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.1 | 0.8 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.8 | GO:0043270 | positive regulation of ion transport(GO:0043270) |
| 0.1 | 0.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.9 | GO:0045116 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 1.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 1.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.1 | 1.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.3 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 3.9 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.1 | 0.4 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 1.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 2.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 6.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.9 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.8 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.8 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 4.1 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.9 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 2.3 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.6 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 4.2 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.9 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.4 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.2 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 2.6 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.0 | 1.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 1.6 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 2.2 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.9 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 3.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.1 | GO:0072657 | protein localization to membrane(GO:0072657) |
| 0.0 | 0.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 1.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 4.9 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.2 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.8 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.4 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 1.6 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 1.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 6.2 | GO:0006886 | intracellular protein transport(GO:0006886) |
| 0.0 | 3.1 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.8 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.6 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 1.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 1.3 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.3 | GO:0097576 | autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.0 | 1.1 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 1.4 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 0.0 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.6 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.1 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.5 | 4.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 1.4 | 13.9 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 1.3 | 7.9 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 1.3 | 11.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 1.2 | 17.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 1.0 | 3.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.9 | 3.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.9 | 2.7 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.9 | 2.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.8 | 3.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.7 | 2.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.7 | 2.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.6 | 3.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 1.7 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.6 | 1.7 | GO:0008352 | katanin complex(GO:0008352) |
| 0.5 | 1.6 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.5 | 2.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.5 | 2.5 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.5 | 7.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 1.6 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.4 | 1.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 1.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.4 | 1.8 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.3 | 4.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 1.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 2.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.3 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 6.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.3 | 5.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 1.2 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.3 | 4.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 1.9 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 7.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 0.8 | GO:0072380 | TRC complex(GO:0072380) |
| 0.3 | 1.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 1.0 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.3 | 6.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 2.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 0.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 1.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 2.1 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.2 | 3.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.2 | 0.6 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.2 | 1.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.2 | 1.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 1.7 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.2 | 4.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 1.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 1.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 2.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.7 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 18.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.2 | 1.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 0.5 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.2 | 0.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 5.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.3 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 2.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 2.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 6.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 4.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial intermembrane space protein transporter complex(GO:0042719) mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.8 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 22.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 1.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 2.0 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 9.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 13.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.9 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 1.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 4.9 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 8.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.3 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 4.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 2.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 3.4 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 3.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 6.1 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.1 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 2.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.0 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.1 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 4.7 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.0 | 3.0 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 5.9 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 6.3 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 14.4 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 2.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.9 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 2.1 | 8.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 2.1 | 6.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 1.6 | 4.9 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.4 | 4.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 1.3 | 5.0 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 1.2 | 3.6 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 1.0 | 4.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 1.0 | 3.9 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 1.0 | 2.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 1.0 | 4.8 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 1.0 | 26.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.9 | 3.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.9 | 2.7 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.9 | 6.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.9 | 2.6 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.8 | 3.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.8 | 2.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.8 | 4.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.7 | 2.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.7 | 2.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.7 | 4.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.7 | 13.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.7 | 2.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.6 | 1.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.6 | 2.9 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.6 | 1.7 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.5 | 3.3 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.5 | 2.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.5 | 5.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.5 | 2.0 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.5 | 5.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.5 | 3.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 6.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.4 | 2.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.4 | 1.3 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.4 | 2.6 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.4 | 5.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 2.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.4 | 1.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 1.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 1.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 2.8 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.4 | 2.0 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.4 | 5.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 1.9 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.4 | 1.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 3.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.4 | 1.5 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.4 | 8.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 2.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.0 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.3 | 5.4 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.3 | 1.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 2.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 0.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.3 | 0.9 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.3 | 1.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 3.2 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 3.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 1.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 0.8 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.3 | 3.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 2.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 8.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.3 | 3.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 1.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 6.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 0.8 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 1.0 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.2 | 2.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 5.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.9 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.2 | 4.0 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.2 | 2.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 3.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 1.6 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.2 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 7.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 3.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.2 | 1.7 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.2 | 1.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 1.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 2.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 3.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 1.6 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 2.7 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 2.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.2 | 1.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 2.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.2 | 1.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.2 | 0.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 4.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 1.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.2 | 1.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 2.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 0.5 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.2 | 1.9 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.9 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 2.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 5.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 2.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 2.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 0.5 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.2 | 1.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 1.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 3.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 2.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 1.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.9 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 4.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.7 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 5.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 5.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 3.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 1.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 0.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 1.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 5.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.9 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.9 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.5 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 4.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.8 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 4.8 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 2.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 5.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.6 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.1 | 1.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.6 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 2.1 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 0.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 5.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 4.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.0 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
| 0.1 | 1.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.3 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.1 | 1.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 2.8 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 4.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.6 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 3.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.9 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 2.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 4.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 4.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 8.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 2.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 3.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 4.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 2.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 1.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 3.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 11.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.6 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 3.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 1.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.2 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.2 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 6.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 2.9 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 4.9 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 2.2 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 1.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 5.4 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 6.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 11.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 3.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 2.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 3.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 4.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 4.1 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 3.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 1.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 6.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 4.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 1.3 | 14.1 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.6 | 2.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 6.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.5 | 4.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 9.6 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.5 | 6.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 6.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 1.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 1.6 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.4 | 7.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 3.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 2.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 6.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 5.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 10.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.3 | 2.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 3.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 2.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 1.9 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 1.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 2.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 2.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 3.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 2.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 1.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 4.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 1.6 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 2.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 1.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 2.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 5.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 2.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 3.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.4 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 2.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 3.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 8.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.6 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 3.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 3.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |