Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
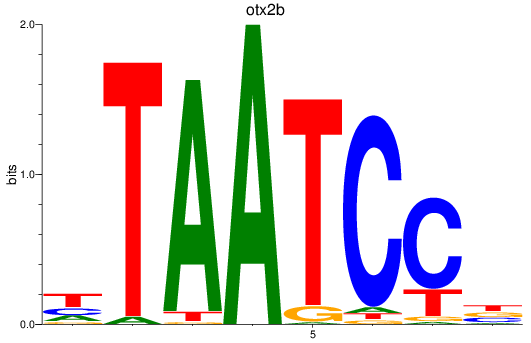
Results for otx2b
Z-value: 1.07
Transcription factors associated with otx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
otx2b
|
ENSDARG00000011235 | orthodenticle homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| otx2b | dr11_v1_chr17_-_44249538_44249538 | 0.75 | 3.7e-18 | Click! |
Activity profile of otx2b motif
Sorted Z-values of otx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_5103349 | 21.18 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr16_+_32059785 | 17.88 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr2_-_30784198 | 16.31 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr14_-_36378494 | 15.62 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr17_-_12389259 | 12.62 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr3_+_34919810 | 12.54 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr11_-_29082429 | 12.53 |
ENSDART00000041443
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr14_-_4273396 | 11.78 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr15_+_33070939 | 11.43 |
ENSDART00000164928
|
mab21l1
|
mab-21-like 1 |
| chr11_-_29082175 | 11.42 |
ENSDART00000123245
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr11_+_39672874 | 10.09 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr3_+_34220194 | 9.70 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr6_+_36942966 | 9.26 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr12_-_10275320 | 9.14 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr5_-_28625515 | 9.05 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
| chr7_-_49594995 | 8.67 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr9_+_13714379 | 8.43 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr3_+_15296824 | 7.87 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr17_-_20979077 | 7.71 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr6_-_20875111 | 7.70 |
ENSDART00000115118
ENSDART00000159916 |
tns1a
|
tensin 1a |
| chr16_-_33059246 | 7.51 |
ENSDART00000171718
ENSDART00000168305 ENSDART00000166401 |
snap91
|
synaptosomal-associated protein 91 |
| chr14_+_34966598 | 7.48 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr15_-_26636826 | 7.45 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr7_-_52842605 | 7.36 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr3_+_1492174 | 7.10 |
ENSDART00000112979
|
sox10
|
SRY (sex determining region Y)-box 10 |
| chr19_-_9503473 | 7.01 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr17_+_18117029 | 6.75 |
ENSDART00000154646
ENSDART00000179739 |
bcl11ba
|
B cell CLL/lymphoma 11Ba |
| chr12_+_15622621 | 6.66 |
ENSDART00000079784
|
plcd3b
|
phospholipase C, delta 3b |
| chr2_+_28995776 | 6.64 |
ENSDART00000138733
|
cdh12a
|
cadherin 12, type 2a (N-cadherin 2) |
| chr10_-_21054059 | 6.50 |
ENSDART00000139733
|
pcdh1a
|
protocadherin 1a |
| chr25_-_19090479 | 6.39 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr20_-_20821783 | 6.29 |
ENSDART00000152577
ENSDART00000027603 ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr15_-_18574716 | 6.20 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr17_+_35097024 | 6.17 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr5_+_43072951 | 6.12 |
ENSDART00000193905
ENSDART00000026020 |
wdr54
|
WD repeat domain 54 |
| chr2_-_4797512 | 6.06 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr5_-_28606916 | 6.05 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr21_+_32339158 | 6.04 |
ENSDART00000161723
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr21_+_32338897 | 5.92 |
ENSDART00000110137
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr20_-_9963713 | 5.83 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr21_-_40782393 | 5.82 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr12_+_25640480 | 5.78 |
ENSDART00000105608
|
prkcea
|
protein kinase C, epsilon a |
| chr2_+_25929619 | 5.74 |
ENSDART00000137746
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr19_-_8880688 | 5.52 |
ENSDART00000039629
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr1_-_16507812 | 5.42 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr23_-_7125494 | 5.33 |
ENSDART00000111929
|
slco4a1
|
solute carrier organic anion transporter family, member 4A1 |
| chr12_+_41991635 | 5.26 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr25_-_7925269 | 5.18 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr25_-_7925019 | 5.12 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr3_-_32170850 | 5.10 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr11_+_25010491 | 5.09 |
ENSDART00000167285
|
zgc:92107
|
zgc:92107 |
| chr5_-_14390445 | 5.02 |
ENSDART00000026120
|
ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr24_+_24923166 | 4.98 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr3_-_28665291 | 4.90 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr11_-_29563437 | 4.88 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr17_+_25289431 | 4.76 |
ENSDART00000161002
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr13_+_1089942 | 4.66 |
ENSDART00000054322
|
cnrip1b
|
cannabinoid receptor interacting protein 1b |
| chr14_-_32503363 | 4.60 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr2_-_1569250 | 4.58 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr1_-_20273227 | 4.39 |
ENSDART00000146084
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr11_-_27702778 | 4.38 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr2_-_48171441 | 4.29 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr7_+_30970045 | 4.24 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr13_-_36525982 | 4.12 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr16_-_37372613 | 4.11 |
ENSDART00000124090
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr18_+_7639401 | 4.11 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr7_-_18656069 | 3.99 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr16_+_11151699 | 3.99 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr12_+_39685485 | 3.89 |
ENSDART00000163403
|
LO017650.1
|
|
| chr20_-_16156419 | 3.88 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr6_+_32415132 | 3.87 |
ENSDART00000155790
|
kank4
|
KN motif and ankyrin repeat domains 4 |
| chr17_-_31058900 | 3.86 |
ENSDART00000134998
ENSDART00000104307 ENSDART00000172721 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr5_+_59494079 | 3.84 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr2_-_16217344 | 3.84 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_-_64770456 | 3.67 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr7_-_41489822 | 3.66 |
ENSDART00000006724
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr10_-_7988396 | 3.63 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr5_+_34981584 | 3.61 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr16_+_24684107 | 3.36 |
ENSDART00000183920
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr5_-_27994679 | 3.25 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr19_+_2631565 | 3.21 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr11_-_4235811 | 3.18 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr9_-_18568927 | 3.17 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr3_-_33574576 | 3.16 |
ENSDART00000184881
|
CR847537.1
|
|
| chr7_-_41489997 | 3.10 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr2_+_11923824 | 3.02 |
ENSDART00000153873
|
trove2
|
TROVE domain family, member 2 |
| chr17_+_10593398 | 3.01 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr8_-_25566347 | 2.95 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr18_+_49969568 | 2.92 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr16_+_42829735 | 2.90 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr15_-_15469079 | 2.87 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr18_+_10784730 | 2.87 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr10_+_29963518 | 2.85 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr10_+_4095917 | 2.80 |
ENSDART00000114533
|
mn1a
|
meningioma 1a |
| chr25_-_16818195 | 2.80 |
ENSDART00000185215
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr4_-_9173552 | 2.75 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr13_+_17468161 | 2.71 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr5_+_37290260 | 2.66 |
ENSDART00000113642
|
zgc:163098
|
zgc:163098 |
| chr7_+_38395197 | 2.66 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr16_+_42830152 | 2.57 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr17_-_50010121 | 2.56 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr9_+_26103814 | 2.52 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr25_+_33002963 | 2.49 |
ENSDART00000187366
|
CABZ01046980.1
|
|
| chr7_+_39011355 | 2.46 |
ENSDART00000173855
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr1_-_39909985 | 2.44 |
ENSDART00000181673
|
stox2a
|
storkhead box 2a |
| chr12_+_11080776 | 2.43 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr5_+_37290586 | 2.43 |
ENSDART00000122919
|
zgc:163098
|
zgc:163098 |
| chr15_+_37559570 | 2.41 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr1_-_17650223 | 2.38 |
ENSDART00000043484
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr23_-_28294763 | 2.32 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr1_+_33383644 | 2.25 |
ENSDART00000187194
|
dhrsx
|
dehydrogenase/reductase (SDR family) X-linked |
| chr13_-_36184701 | 2.21 |
ENSDART00000146522
|
map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr14_-_30587814 | 2.19 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr24_-_1303935 | 2.19 |
ENSDART00000159212
ENSDART00000159267 ENSDART00000164904 |
nrp1a
|
neuropilin 1a |
| chr3_-_61593274 | 2.14 |
ENSDART00000154132
ENSDART00000055071 |
nptx2a
|
neuronal pentraxin 2a |
| chr14_-_46395408 | 2.10 |
ENSDART00000147537
|
bbs7
|
Bardet-Biedl syndrome 7 |
| chr16_+_14710436 | 2.01 |
ENSDART00000027982
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr7_-_12821277 | 1.99 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr4_+_1722899 | 1.97 |
ENSDART00000021139
|
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr1_-_26293203 | 1.95 |
ENSDART00000180140
|
cxxc4
|
CXXC finger 4 |
| chr14_-_29799993 | 1.92 |
ENSDART00000133775
ENSDART00000005568 |
pdlim3b
|
PDZ and LIM domain 3b |
| chr9_-_32343673 | 1.88 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr3_-_30885250 | 1.80 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
| chr15_-_15468326 | 1.75 |
ENSDART00000161192
|
rab34a
|
RAB34, member RAS oncogene family a |
| chr25_-_16818380 | 1.63 |
ENSDART00000155401
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr8_+_14381272 | 1.62 |
ENSDART00000057642
|
acbd6
|
acyl-CoA binding domain containing 6 |
| chr1_-_11075403 | 1.61 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr22_+_30184039 | 1.60 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr11_-_3860199 | 1.58 |
ENSDART00000082420
|
gata2a
|
GATA binding protein 2a |
| chr10_-_25561751 | 1.55 |
ENSDART00000147089
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chrM_+_15308 | 1.49 |
ENSDART00000093625
|
mt-cyb
|
cytochrome b, mitochondrial |
| chr3_-_28828242 | 1.48 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr25_-_18913336 | 1.47 |
ENSDART00000171010
|
parietopsin
|
parietopsin |
| chr18_+_5172848 | 1.45 |
ENSDART00000190642
|
CABZ01080599.1
|
|
| chr25_+_29688671 | 1.37 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr14_-_32089117 | 1.36 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr25_-_35996141 | 1.36 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr5_-_17876709 | 1.35 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr21_-_28235361 | 1.32 |
ENSDART00000164458
|
nrxn2a
|
neurexin 2a |
| chr2_-_48171112 | 1.30 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr14_+_14847745 | 1.29 |
ENSDART00000189302
ENSDART00000188336 |
ccdc142
|
coiled-coil domain containing 142 |
| chr19_+_43884120 | 1.29 |
ENSDART00000139684
ENSDART00000142312 |
lypla2
|
lysophospholipase II |
| chr6_-_17999776 | 1.27 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr13_+_31205439 | 1.26 |
ENSDART00000132326
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr11_+_2506516 | 1.22 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr15_+_29116063 | 1.18 |
ENSDART00000016112
ENSDART00000153609 ENSDART00000155630 |
capns1b
|
calpain, small subunit 1 b |
| chr12_+_17042754 | 1.10 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr10_-_18468385 | 1.10 |
ENSDART00000183302
|
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr9_-_8454060 | 1.06 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr5_-_38107741 | 1.04 |
ENSDART00000156853
|
si:ch211-284e13.14
|
si:ch211-284e13.14 |
| chr7_+_57866292 | 1.04 |
ENSDART00000138757
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr24_-_18877118 | 0.97 |
ENSDART00000092783
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr20_-_16548912 | 0.97 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr14_-_2400668 | 0.96 |
ENSDART00000172717
ENSDART00000182882 |
si:ch73-233f7.1
|
si:ch73-233f7.1 |
| chr24_+_3478871 | 0.94 |
ENSDART00000111491
ENSDART00000134598 ENSDART00000142407 |
wdr37
|
WD repeat domain 37 |
| chr11_-_40257225 | 0.94 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr9_-_48184823 | 0.93 |
ENSDART00000180264
|
klhl23
|
kelch-like family member 23 |
| chr14_+_14847304 | 0.93 |
ENSDART00000169932
|
ccdc142
|
coiled-coil domain containing 142 |
| chr20_-_34768301 | 0.92 |
ENSDART00000142482
|
paqr8
|
progestin and adipoQ receptor family member VIII |
| chr20_+_16721933 | 0.91 |
ENSDART00000063950
|
psmc1b
|
proteasome 26S subunit, ATPase 1b |
| chr24_-_18876877 | 0.88 |
ENSDART00000186269
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr21_-_41369370 | 0.88 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr4_-_5077158 | 0.86 |
ENSDART00000155915
|
ahcyl2
|
adenosylhomocysteinase-like 2 |
| chr7_+_39011852 | 0.82 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr14_-_2933185 | 0.81 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr11_-_16215143 | 0.76 |
ENSDART00000027014
|
rab7
|
RAB7, member RAS oncogene family |
| chr3_+_23703704 | 0.74 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr13_-_47800342 | 0.73 |
ENSDART00000121817
|
fbln7
|
fibulin 7 |
| chr9_+_11034967 | 0.72 |
ENSDART00000152081
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr2_+_31476065 | 0.72 |
ENSDART00000049219
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr16_+_29376751 | 0.71 |
ENSDART00000168856
ENSDART00000162502 ENSDART00000050535 |
rrnad1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr25_+_19753890 | 0.69 |
ENSDART00000190268
ENSDART00000128852 |
celsr1b
|
cadherin EGF LAG seven-pass G-type receptor 1b |
| chr23_-_27050083 | 0.68 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr22_-_35006554 | 0.67 |
ENSDART00000138639
|
kcnip2
|
Kv channel interacting protein 2 |
| chr13_-_50614639 | 0.67 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr16_+_16265850 | 0.66 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr16_+_3067134 | 0.65 |
ENSDART00000012048
|
cyb5r4
|
cytochrome b5 reductase 4 |
| chr11_-_3860534 | 0.64 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr2_+_30878864 | 0.63 |
ENSDART00000009326
|
oprk1
|
opioid receptor, kappa 1 |
| chr18_+_19006063 | 0.60 |
ENSDART00000135729
|
slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr2_-_50272077 | 0.58 |
ENSDART00000127623
|
cul1a
|
cullin 1a |
| chr23_-_3759692 | 0.55 |
ENSDART00000028885
|
hmga1a
|
high mobility group AT-hook 1a |
| chr1_+_44523516 | 0.37 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr14_-_25034142 | 0.34 |
ENSDART00000000392
|
pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr5_-_14344647 | 0.33 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr10_+_9222476 | 0.32 |
ENSDART00000064973
ENSDART00000145879 |
paqr3b
|
progestin and adipoQ receptor family member IIIb |
| chr7_-_35516251 | 0.30 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr12_-_35582521 | 0.28 |
ENSDART00000162175
ENSDART00000168958 |
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr7_+_66884291 | 0.24 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr18_-_19005919 | 0.24 |
ENSDART00000129776
|
ints14
|
integrator complex subunit 14 |
| chr2_-_9744081 | 0.18 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr17_-_21617618 | 0.13 |
ENSDART00000109031
|
gpr26
|
G protein-coupled receptor 26 |
| chr6_-_36795111 | 0.12 |
ENSDART00000160669
ENSDART00000104256 ENSDART00000187751 ENSDART00000161928 ENSDART00000183264 |
opa1
|
optic atrophy 1 (autosomal dominant) |
| chr17_+_16564921 | 0.12 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr12_-_35582683 | 0.11 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr7_-_17780048 | 0.11 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr20_-_34754617 | 0.10 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr14_+_21754521 | 0.09 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr16_+_37876779 | 0.09 |
ENSDART00000140148
|
si:ch211-198c19.1
|
si:ch211-198c19.1 |
| chr2_-_38337122 | 0.09 |
ENSDART00000076523
ENSDART00000187473 |
slc7a8b
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8b |
Network of associatons between targets according to the STRING database.
First level regulatory network of otx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 2.0 | 12.0 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 1.9 | 21.2 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 1.5 | 4.4 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 1.2 | 14.7 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.9 | 3.6 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.9 | 12.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.8 | 6.8 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.7 | 2.2 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.7 | 5.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 24.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.6 | 3.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.6 | 1.8 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.6 | 4.6 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.6 | 4.6 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.5 | 7.1 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.5 | 4.7 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.5 | 5.6 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.5 | 2.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.5 | 6.3 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 2.2 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.4 | 4.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 6.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.4 | 4.9 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.4 | 3.9 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 5.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 7.4 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.3 | 5.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 7.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 12.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 8.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 0.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 2.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.2 | 6.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 5.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 2.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 1.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 2.9 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 3.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.6 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 4.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.7 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 5.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 2.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 1.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 3.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 6.7 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 2.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.7 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 2.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 5.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 2.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 8.4 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.1 | 4.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.3 | GO:0032656 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 0.7 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 1.3 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 3.9 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 1.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 3.2 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 2.2 | GO:0046854 | lipid phosphorylation(GO:0046834) phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.1 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 4.8 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 2.1 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 2.2 | GO:0048596 | embryonic camera-type eye morphogenesis(GO:0048596) |
| 0.0 | 1.1 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 3.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 1.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 4.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 13.3 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.0 | 4.0 | GO:0007015 | actin filament organization(GO:0007015) |
| 0.0 | 3.9 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 1.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.6 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.3 | 15.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.7 | 7.5 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.5 | 3.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 5.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 5.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.4 | 6.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 2.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 2.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 5.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 5.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 1.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 7.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 5.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 20.9 | GO:0005912 | adherens junction(GO:0005912) |
| 0.1 | 1.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 5.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 5.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 4.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 7.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 5.7 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 3.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 9.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) apical junction complex(GO:0043296) occluding junction(GO:0070160) |
| 0.0 | 9.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 8.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 4.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 5.8 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 13.3 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 0.9 | GO:0000502 | proteasome complex(GO:0000502) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 1.9 | 21.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 1.2 | 5.8 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 1.0 | 5.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.6 | 1.8 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.6 | 7.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.5 | 2.7 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.5 | 4.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.5 | 3.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 5.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.5 | 6.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.5 | 7.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.4 | 12.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.4 | 8.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 12.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 1.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 4.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 5.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 10.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 5.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 5.8 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 5.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.2 | 1.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 6.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.2 | 9.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.2 | 2.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 3.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 0.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 3.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 11.4 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.1 | 1.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 7.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 3.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 4.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.6 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 2.7 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 2.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 12.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 18.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 13.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 11.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 12.5 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 3.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 3.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 8.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.4 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 5.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 12.9 | GO:0008270 | zinc ion binding(GO:0008270) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 16.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.2 | 3.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 16.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.3 | 4.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 3.7 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.2 | 6.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 6.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 2.0 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 3.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |