Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
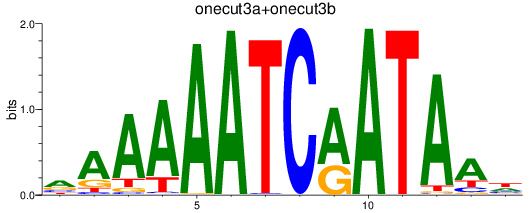
Results for onecut3a+onecut3b
Z-value: 1.18
Transcription factors associated with onecut3a+onecut3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
onecut3a
|
ENSDARG00000056395 | one cut homeobox 3a |
|
onecut3b
|
ENSDARG00000091059 | one cut homeobox 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| onecut3a | dr11_v1_chr22_+_20461488_20461488 | 0.37 | 2.0e-04 | Click! |
Activity profile of onecut3a+onecut3b motif
Sorted Z-values of onecut3a+onecut3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_35748385 | 18.87 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr14_+_35748206 | 18.71 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr1_-_21409877 | 18.47 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr19_+_25649626 | 17.47 |
ENSDART00000146947
|
tac1
|
tachykinin 1 |
| chr6_+_3828560 | 16.10 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr19_-_17526735 | 15.15 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr2_+_47581997 | 14.45 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr16_-_12173554 | 13.93 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr8_-_14052349 | 13.47 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr4_-_5291256 | 13.24 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr3_-_28665291 | 12.63 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr10_-_31782616 | 11.81 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr1_+_16144615 | 11.49 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr18_+_642889 | 11.48 |
ENSDART00000189007
|
CABZ01078320.1
|
|
| chr11_-_10798021 | 11.15 |
ENSDART00000167112
ENSDART00000179725 ENSDART00000091923 ENSDART00000185825 |
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr25_-_11088839 | 10.01 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr5_-_71460556 | 9.89 |
ENSDART00000108804
|
brinp1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr2_-_39759059 | 9.70 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr24_-_23320223 | 9.26 |
ENSDART00000135846
|
zfhx4
|
zinc finger homeobox 4 |
| chr17_-_20897250 | 8.81 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr14_+_21783229 | 8.79 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr2_+_5887626 | 8.12 |
ENSDART00000147831
|
si:ch211-168b3.1
|
si:ch211-168b3.1 |
| chr20_-_30931139 | 7.97 |
ENSDART00000006778
ENSDART00000146376 |
acat2
|
acetyl-CoA acetyltransferase 2 |
| chr17_+_23937262 | 7.87 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr10_+_10801719 | 7.86 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr21_-_17482465 | 7.73 |
ENSDART00000004548
|
barhl1b
|
BarH-like homeobox 1b |
| chr10_-_15405564 | 7.67 |
ENSDART00000020665
|
sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr1_-_23157583 | 7.49 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr11_+_6819050 | 7.32 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr5_+_19309877 | 7.27 |
ENSDART00000190338
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr23_+_12454542 | 7.25 |
ENSDART00000182259
ENSDART00000193043 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr11_+_25064519 | 6.89 |
ENSDART00000016181
|
ndrg3a
|
ndrg family member 3a |
| chr3_+_39853788 | 6.88 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr16_-_12173399 | 6.58 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr15_-_34418525 | 6.47 |
ENSDART00000147582
|
agmo
|
alkylglycerol monooxygenase |
| chr2_+_47582488 | 6.41 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr25_-_9805269 | 6.41 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr3_-_28120092 | 6.24 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr15_+_17441734 | 5.97 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr23_-_28025943 | 5.79 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr21_+_3901775 | 5.79 |
ENSDART00000053609
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr6_-_17999776 | 5.75 |
ENSDART00000183048
ENSDART00000181577 ENSDART00000170597 |
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr18_-_20560007 | 5.70 |
ENSDART00000141367
ENSDART00000090186 |
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr13_+_36144341 | 5.67 |
ENSDART00000182930
ENSDART00000187327 |
TTC9
|
si:ch211-259k16.3 |
| chr18_-_44611252 | 5.37 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr21_-_43482426 | 5.33 |
ENSDART00000192901
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr2_-_56387041 | 5.01 |
ENSDART00000036240
|
cers4b
|
ceramide synthase 4b |
| chr21_-_23475361 | 5.00 |
ENSDART00000156658
ENSDART00000157454 |
ncam1a
|
neural cell adhesion molecule 1a |
| chr2_+_42871831 | 4.99 |
ENSDART00000171393
|
efr3a
|
EFR3 homolog A (S. cerevisiae) |
| chr2_-_32505091 | 4.89 |
ENSDART00000141884
ENSDART00000056639 |
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr7_-_49651378 | 4.86 |
ENSDART00000015040
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr4_-_4261673 | 4.78 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr9_+_32358514 | 4.75 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr4_+_7827261 | 4.62 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr22_+_17359346 | 4.60 |
ENSDART00000145434
|
gpr52
|
G protein-coupled receptor 52 |
| chr23_+_28770225 | 4.55 |
ENSDART00000132179
ENSDART00000142273 |
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr1_-_36151377 | 4.49 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr7_+_63325819 | 4.41 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr14_+_36628131 | 4.36 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr19_-_18855513 | 4.33 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr2_+_44781185 | 4.30 |
ENSDART00000187103
ENSDART00000147292 ENSDART00000098123 |
ece2a
|
endothelin converting enzyme 2a |
| chr5_+_50788329 | 4.25 |
ENSDART00000050979
|
fam81b
|
family with sequence similarity 81, member B |
| chr17_-_8169774 | 4.16 |
ENSDART00000091828
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr19_-_18855717 | 4.04 |
ENSDART00000158192
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr23_-_15216654 | 4.04 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr14_-_4044545 | 3.96 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr24_-_15131831 | 3.93 |
ENSDART00000028410
|
cd226
|
CD226 molecule |
| chr2_+_25860344 | 3.88 |
ENSDART00000178841
|
slc7a14a
|
solute carrier family 7, member 14a |
| chr7_+_40658003 | 3.87 |
ENSDART00000172435
|
lmbr1
|
limb development membrane protein 1 |
| chr3_+_32443395 | 3.83 |
ENSDART00000188447
|
prr12b
|
proline rich 12b |
| chr3_-_30434016 | 3.71 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr11_+_5817202 | 3.67 |
ENSDART00000126084
|
ctdspl3
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 3 |
| chr2_+_21309272 | 3.65 |
ENSDART00000141322
|
zbtb47a
|
zinc finger and BTB domain containing 47a |
| chr20_-_3403033 | 3.63 |
ENSDART00000092264
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr10_-_28835771 | 3.61 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
| chr15_+_17446796 | 3.52 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr6_-_58764672 | 3.44 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr9_+_19421841 | 3.43 |
ENSDART00000159090
ENSDART00000084771 |
pde9a
|
phosphodiesterase 9A |
| chr19_-_21766461 | 3.39 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr12_+_10116912 | 3.39 |
ENSDART00000189630
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr12_+_32729470 | 3.28 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr11_+_24815667 | 3.24 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr8_+_9866351 | 3.19 |
ENSDART00000133985
|
kcnd1
|
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr14_+_11946395 | 3.18 |
ENSDART00000193290
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr17_-_20897407 | 3.17 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr15_+_11381532 | 3.10 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr21_-_39058490 | 2.99 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr3_+_1492174 | 2.95 |
ENSDART00000112979
|
sox10
|
SRY (sex determining region Y)-box 10 |
| chr10_+_7593185 | 2.86 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr1_-_46663997 | 2.86 |
ENSDART00000134450
|
ebpl
|
emopamil binding protein-like |
| chr25_-_13789955 | 2.80 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr14_-_31893996 | 2.74 |
ENSDART00000173222
|
gpr101
|
G protein-coupled receptor 101 |
| chr21_-_41369370 | 2.71 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr11_+_25259058 | 2.68 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr17_-_45247332 | 2.61 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr6_-_40352215 | 2.47 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr22_-_3564563 | 2.46 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr12_-_43428542 | 2.39 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr16_-_44127307 | 2.35 |
ENSDART00000104583
ENSDART00000151936 ENSDART00000058685 ENSDART00000190830 |
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr3_-_48612078 | 2.35 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr11_+_24815927 | 2.34 |
ENSDART00000146838
|
rabif
|
RAB interacting factor |
| chr16_-_54942532 | 2.25 |
ENSDART00000078887
ENSDART00000101402 |
tmem222a
|
transmembrane protein 222a |
| chr23_+_25832689 | 2.19 |
ENSDART00000138907
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr7_+_73649686 | 2.16 |
ENSDART00000185589
|
PCP4L1
|
si:dkey-46i9.1 |
| chr20_-_23171430 | 2.14 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr24_-_38113208 | 2.11 |
ENSDART00000079003
|
crp
|
c-reactive protein, pentraxin-related |
| chr1_+_51391082 | 2.06 |
ENSDART00000063936
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr9_-_20853439 | 2.04 |
ENSDART00000028247
ENSDART00000133321 |
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr10_-_11840353 | 2.02 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr6_-_16456093 | 2.00 |
ENSDART00000083305
ENSDART00000181640 |
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr16_-_26435431 | 1.98 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr12_-_47793857 | 1.94 |
ENSDART00000161294
|
dydc2
|
DPY30 domain containing 2 |
| chr16_+_21738194 | 1.93 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr9_+_27329640 | 1.93 |
ENSDART00000111039
|
GTPBP8
|
si:rp71-84d19.3 |
| chr13_-_37383625 | 1.91 |
ENSDART00000192888
ENSDART00000100322 ENSDART00000138934 |
kcnh5b
|
potassium voltage-gated channel, subfamily H (eag-related), member 5b |
| chr17_-_14451405 | 1.91 |
ENSDART00000146786
|
jkamp
|
jnk1/mapk8 associated membrane protein |
| chr2_-_6112862 | 1.86 |
ENSDART00000164269
|
prdx1
|
peroxiredoxin 1 |
| chr8_-_23081511 | 1.85 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr12_+_48204891 | 1.85 |
ENSDART00000190534
ENSDART00000164427 |
ndr2
|
nodal-related 2 |
| chr7_-_24022340 | 1.83 |
ENSDART00000149133
|
cideb
|
cell death-inducing DFFA-like effector b |
| chr7_-_56831621 | 1.81 |
ENSDART00000182912
|
senp8
|
SUMO/sentrin peptidase family member, NEDD8 specific |
| chr13_-_31166544 | 1.81 |
ENSDART00000146250
ENSDART00000132129 ENSDART00000139591 |
mapk8a
|
mitogen-activated protein kinase 8a |
| chr13_-_4664403 | 1.81 |
ENSDART00000023803
ENSDART00000177957 |
c1d
|
C1D nuclear receptor corepressor |
| chr4_+_27018663 | 1.80 |
ENSDART00000180778
|
CR749777.1
|
|
| chr22_-_38360205 | 1.72 |
ENSDART00000162055
|
mark1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr4_+_8016457 | 1.70 |
ENSDART00000014036
|
optn
|
optineurin |
| chr20_+_23440632 | 1.64 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr3_+_39958407 | 1.64 |
ENSDART00000099754
|
ccdc97
|
coiled-coil domain containing 97 |
| chr8_-_14484599 | 1.60 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr18_+_3579829 | 1.52 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr1_+_37391716 | 1.51 |
ENSDART00000191986
|
sparcl1
|
SPARC-like 1 |
| chr4_-_76488854 | 1.50 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr13_-_30713236 | 1.49 |
ENSDART00000112372
ENSDART00000142221 |
tmem72
|
transmembrane protein 72 |
| chr5_-_57652168 | 1.49 |
ENSDART00000166245
|
gb:eh456644
|
expressed sequence EH456644 |
| chr18_+_15876385 | 1.48 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr4_+_25691642 | 1.42 |
ENSDART00000139053
|
acot18
|
acyl-CoA thioesterase 18 |
| chr9_+_38503233 | 1.39 |
ENSDART00000140331
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr10_-_28761454 | 1.38 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr2_+_45300512 | 1.33 |
ENSDART00000144704
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr12_+_47794089 | 1.30 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr8_+_40477264 | 1.27 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr5_+_56268436 | 1.25 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr17_-_26507289 | 1.24 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr14_+_49376011 | 1.23 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr6_+_40992409 | 1.22 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr3_-_60142530 | 1.21 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr24_+_23730061 | 1.15 |
ENSDART00000080343
|
ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr13_+_49727333 | 1.15 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr20_+_16639848 | 1.13 |
ENSDART00000063944
ENSDART00000152359 |
tmem30ab
|
transmembrane protein 30Ab |
| chr5_+_58455488 | 1.12 |
ENSDART00000038602
ENSDART00000127958 |
slc37a2
|
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr11_+_37201483 | 1.11 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr19_+_11984725 | 1.10 |
ENSDART00000185960
|
spag1a
|
sperm associated antigen 1a |
| chr21_+_6212844 | 1.06 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr7_-_68419830 | 0.99 |
ENSDART00000191606
|
CU468723.2
|
|
| chr22_-_7669624 | 0.98 |
ENSDART00000189617
|
AL929276.2
|
|
| chr5_+_3118231 | 0.96 |
ENSDART00000179809
|
supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr18_-_44847855 | 0.96 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr7_-_41693004 | 0.91 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr16_-_26255877 | 0.91 |
ENSDART00000146214
|
erfl1
|
Ets2 repressor factor like 1 |
| chr2_+_13462305 | 0.90 |
ENSDART00000149309
ENSDART00000080900 |
cfap57
|
cilia and flagella associated protein 57 |
| chr12_-_35582683 | 0.89 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr2_-_17044959 | 0.87 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr1_+_47391404 | 0.86 |
ENSDART00000113559
|
bcl9
|
B cell CLL/lymphoma 9 |
| chr1_+_12177195 | 0.82 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr25_+_22017182 | 0.76 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr3_-_58165254 | 0.75 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr19_+_43256986 | 0.74 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr6_-_33707278 | 0.68 |
ENSDART00000188103
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr7_-_36096582 | 0.68 |
ENSDART00000188507
|
CR848715.1
|
|
| chr16_-_2650341 | 0.68 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr15_+_21947117 | 0.68 |
ENSDART00000156234
|
ttc12
|
tetratricopeptide repeat domain 12 |
| chr20_+_42978499 | 0.67 |
ENSDART00000138793
|
si:ch211-203k16.3
|
si:ch211-203k16.3 |
| chr11_+_691734 | 0.67 |
ENSDART00000191463
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr16_+_23960744 | 0.64 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr3_+_24050043 | 0.62 |
ENSDART00000151788
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr8_+_12925385 | 0.57 |
ENSDART00000085377
|
KIF2A
|
zgc:103670 |
| chr16_+_33655890 | 0.57 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr9_+_907459 | 0.54 |
ENSDART00000034850
ENSDART00000144114 |
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr2_-_59145027 | 0.54 |
ENSDART00000128320
|
FO834803.1
|
|
| chr20_-_40367493 | 0.53 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr12_-_18883079 | 0.53 |
ENSDART00000178619
|
shisa8b
|
shisa family member 8b |
| chr22_-_3092136 | 0.53 |
ENSDART00000157620
|
ing5a
|
inhibitor of growth family, member 5a |
| chr22_-_16494406 | 0.50 |
ENSDART00000062727
|
stx6
|
syntaxin 6 |
| chr24_-_25030173 | 0.48 |
ENSDART00000126395
ENSDART00000019350 |
nup58
|
nucleoporin 58 |
| chr15_-_7598542 | 0.47 |
ENSDART00000173092
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr22_-_9839003 | 0.45 |
ENSDART00000193573
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr22_+_9872323 | 0.34 |
ENSDART00000129240
|
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr16_-_47426482 | 0.30 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr10_-_28477023 | 0.28 |
ENSDART00000137008
|
bbx
|
bobby sox homolog (Drosophila) |
| chr15_-_7598294 | 0.20 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr2_-_39086540 | 0.20 |
ENSDART00000110826
|
NMNAT3
|
si:ch73-170l17.1 |
| chr22_+_25554232 | 0.16 |
ENSDART00000131218
|
aars2
|
alanyl-tRNA synthetase 2, mitochondrial (putative) |
| chr19_-_874888 | 0.13 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr3_+_32689707 | 0.12 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr21_+_18877130 | 0.11 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr19_-_48039400 | 0.11 |
ENSDART00000166748
ENSDART00000165921 |
csf3b
|
colony stimulating factor 3 (granulocyte) b |
| chr16_-_26232411 | 0.09 |
ENSDART00000139355
|
arhgef1b
|
Rho guanine nucleotide exchange factor (GEF) 1b |
| chr11_-_22605981 | 0.09 |
ENSDART00000186923
|
myog
|
myogenin |
| chr20_+_34151670 | 0.08 |
ENSDART00000152870
ENSDART00000010329 ENSDART00000145852 |
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr16_+_35728992 | 0.04 |
ENSDART00000158442
|
map7d1a
|
MAP7 domain containing 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of onecut3a+onecut3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 3.8 | 15.2 | GO:0042671 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 3.5 | 17.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 3.2 | 9.7 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 2.8 | 2.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.6 | 7.7 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 2.4 | 7.3 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 2.3 | 20.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 1.7 | 5.0 | GO:0021961 | posterior commissure morphogenesis(GO:0021961) |
| 1.3 | 3.9 | GO:0002347 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 1.3 | 6.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.2 | 18.5 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 1.1 | 3.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.0 | 9.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.9 | 5.4 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.7 | 11.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.7 | 4.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.5 | 4.7 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.5 | 20.9 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.5 | 11.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.4 | 1.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.4 | 6.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.4 | 3.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.4 | 2.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.4 | 3.4 | GO:0034433 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.4 | 1.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 5.4 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.3 | 1.8 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.3 | 4.0 | GO:0010573 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 4.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 5.8 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.2 | 1.9 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.6 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 2.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 2.1 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.2 | 1.6 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 2.0 | GO:0045117 | azole transport(GO:0045117) |
| 0.2 | 8.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 1.9 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 2.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.2 | 5.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.2 | 4.2 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.2 | 0.8 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 7.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 7.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 3.6 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 2.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.7 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 1.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 5.0 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 6.2 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 1.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 1.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 7.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.9 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 7.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.1 | 5.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 1.4 | GO:0048885 | neuromast deposition(GO:0048885) |
| 0.1 | 4.8 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.1 | 2.7 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 5.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 10.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 3.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 3.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 1.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.8 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 2.1 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.8 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 5.0 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.1 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.1 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 1.0 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 5.3 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.5 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 3.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 4.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.4 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.1 | GO:0016125 | sterol metabolic process(GO:0016125) |
| 0.0 | 0.7 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 2.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0072380 | TRC complex(GO:0072380) |
| 1.8 | 7.2 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 1.3 | 56.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.8 | 11.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.7 | 5.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.3 | 1.0 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.3 | 6.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.3 | 3.6 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.2 | 20.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 4.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 2.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 10.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 2.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 9.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.8 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 31.6 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 11.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 17.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 7.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 2.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 5.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 4.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 8.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 6.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 2.5 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 16.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 14.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 7.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.1 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 1.7 | GO:0036477 | dendrite(GO:0030425) somatodendritic compartment(GO:0036477) |
| 0.0 | 0.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.5 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 5.4 | 16.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 3.8 | 15.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 3.7 | 56.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 3.2 | 9.7 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 2.7 | 8.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 1.0 | 6.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.9 | 3.4 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.8 | 2.5 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.8 | 11.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.7 | 6.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.5 | 3.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.5 | 13.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 2.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.4 | 1.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 5.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 3.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 1.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 3.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 1.8 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 2.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.2 | 4.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 4.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.3 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.7 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.2 | 2.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 3.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 4.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 5.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 7.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 4.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 12.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 5.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 10.1 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 2.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 4.7 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 7.7 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 8.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 6.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.5 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 4.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 3.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 3.7 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 10.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.4 | GO:0000149 | SNARE binding(GO:0000149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 15.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 12.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.8 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 5.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.4 | 17.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 5.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 3.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 2.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 3.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 4.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 11.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 11.1 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 3.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 2.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 3.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.5 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |