Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
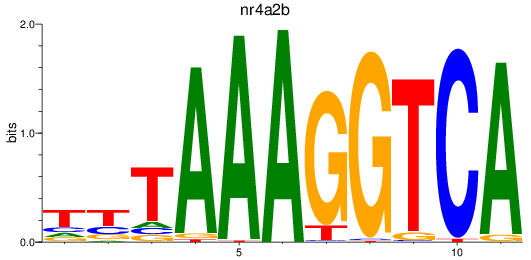
Results for nr4a2b
Z-value: 1.00
Transcription factors associated with nr4a2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a2b
|
ENSDARG00000044532 | nuclear receptor subfamily 4, group A, member 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a2b | dr11_v1_chr6_+_12462079_12462079 | 0.39 | 1.1e-04 | Click! |
Activity profile of nr4a2b motif
Sorted Z-values of nr4a2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_40150612 | 13.97 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr7_+_31871830 | 13.76 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_39764995 | 11.40 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr11_+_11201096 | 11.33 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr12_-_26415499 | 10.85 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr14_-_17121676 | 10.42 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr3_+_39566999 | 10.34 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr6_-_39765546 | 9.55 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr24_+_25471196 | 9.05 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr12_-_26423439 | 8.62 |
ENSDART00000113978
|
synpo2lb
|
synaptopodin 2-like b |
| chr3_+_30257582 | 8.37 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr10_-_17745345 | 7.63 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr2_-_48753873 | 7.13 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr6_-_21189295 | 6.69 |
ENSDART00000137136
|
obsl1a
|
obscurin-like 1a |
| chr5_-_68093169 | 6.60 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr11_+_1409622 | 5.79 |
ENSDART00000157042
|
ppp1r3db
|
protein phosphatase 1, regulatory subunit 3Db |
| chr20_-_10120442 | 5.54 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr6_-_31348999 | 5.28 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr24_+_34089977 | 5.16 |
ENSDART00000157466
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr13_-_41155472 | 5.14 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr7_+_25913225 | 5.10 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr5_+_1278092 | 5.04 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr3_+_31177972 | 4.97 |
ENSDART00000185954
|
clec19a
|
C-type lectin domain containing 19A |
| chr3_-_22212764 | 4.82 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr17_-_12389259 | 4.43 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr5_-_16983336 | 4.39 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr22_+_30184039 | 4.28 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr4_-_4932619 | 4.26 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr9_+_31795343 | 4.02 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr8_-_14052349 | 3.94 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr8_+_13106760 | 3.84 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr3_-_28258462 | 3.82 |
ENSDART00000191573
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr12_+_14084291 | 3.62 |
ENSDART00000189734
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr12_+_46708920 | 3.55 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr21_+_5080789 | 3.52 |
ENSDART00000024199
|
atp5fa1
|
ATP synthase F1 subunit alpha |
| chr6_-_35738836 | 3.51 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr6_-_10835849 | 3.51 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr13_-_21739142 | 3.47 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr3_-_46818001 | 3.45 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr7_+_38588866 | 3.43 |
ENSDART00000015682
|
ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) |
| chr3_-_15154871 | 3.43 |
ENSDART00000147704
|
si:dkey-282h22.5
|
si:dkey-282h22.5 |
| chr2_-_32457919 | 3.31 |
ENSDART00000132792
ENSDART00000041319 |
slc4a2a
|
solute carrier family 4 (anion exchanger), member 2a |
| chr25_-_16969922 | 3.29 |
ENSDART00000111158
|
tigara
|
tp53-induced glycolysis and apoptosis regulator a |
| chr6_+_13206516 | 3.13 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr3_+_23687909 | 3.12 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr11_-_21363834 | 3.10 |
ENSDART00000080051
|
RASSF5
|
si:dkey-85p17.3 |
| chr5_+_36611128 | 3.10 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr9_+_42066030 | 3.08 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr8_-_17997845 | 3.07 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr1_+_16127825 | 2.77 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr9_-_21067971 | 2.77 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr8_+_39634114 | 2.75 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr10_+_41668483 | 2.67 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr14_+_30340251 | 2.67 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr1_-_58868306 | 2.64 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr22_-_11833317 | 2.58 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr3_-_46817838 | 2.55 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr3_+_17806213 | 2.52 |
ENSDART00000055890
|
znf385c
|
zinc finger protein 385C |
| chr7_-_19146925 | 2.52 |
ENSDART00000142924
ENSDART00000009695 |
kirrel1a
|
kirre like nephrin family adhesion molecule 1a |
| chr6_-_10320676 | 2.50 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr18_+_30511230 | 2.46 |
ENSDART00000078888
|
dusp22a
|
dual specificity phosphatase 22a |
| chr15_+_20403903 | 2.43 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr2_+_34767171 | 2.40 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr4_-_16451375 | 2.33 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr17_+_7595356 | 2.21 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr17_+_30545895 | 2.19 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr18_+_9382847 | 2.04 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr18_+_27926839 | 1.99 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr18_+_31410652 | 1.98 |
ENSDART00000098504
|
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr8_-_979735 | 1.97 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr19_+_32158010 | 1.97 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr6_-_29288155 | 1.96 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr7_+_44484853 | 1.96 |
ENSDART00000189079
ENSDART00000121826 |
bean1
|
brain expressed, associated with NEDD4, 1 |
| chr11_+_2172335 | 1.93 |
ENSDART00000170593
|
hoxc12b
|
homeobox C12b |
| chr6_-_50404732 | 1.87 |
ENSDART00000055510
|
romo1
|
reactive oxygen species modulator 1 |
| chr8_-_44004135 | 1.82 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr6_+_29288223 | 1.81 |
ENSDART00000112099
|
zgc:172121
|
zgc:172121 |
| chr12_-_29305220 | 1.73 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr16_+_20161805 | 1.73 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr12_-_47648538 | 1.70 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr18_-_14922872 | 1.70 |
ENSDART00000091722
|
panx2
|
pannexin 2 |
| chr14_+_30413758 | 1.69 |
ENSDART00000092953
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr24_+_22022109 | 1.67 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr9_+_24065855 | 1.66 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr7_+_38667066 | 1.65 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr13_+_21768447 | 1.62 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr1_-_30689004 | 1.61 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr25_+_16214854 | 1.57 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr2_-_11912347 | 1.57 |
ENSDART00000023851
|
abhd3
|
abhydrolase domain containing 3 |
| chr23_+_21638258 | 1.54 |
ENSDART00000104188
|
igsf21b
|
immunoglobin superfamily, member 21b |
| chr12_+_36891483 | 1.52 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr6_+_29288006 | 1.49 |
ENSDART00000043496
|
zgc:172121
|
zgc:172121 |
| chr22_+_31821815 | 1.49 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr1_+_31638274 | 1.47 |
ENSDART00000057885
|
fgf8b
|
fibroblast growth factor 8 b |
| chr17_-_17764801 | 1.44 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr5_-_26330313 | 1.44 |
ENSDART00000148656
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr24_+_684924 | 1.44 |
ENSDART00000186103
|
si:ch211-188f17.1
|
si:ch211-188f17.1 |
| chr24_-_24848612 | 1.41 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr13_-_27660955 | 1.38 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr16_-_25233515 | 1.36 |
ENSDART00000058943
|
zgc:110182
|
zgc:110182 |
| chr15_+_1705167 | 1.34 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr7_-_31794878 | 1.33 |
ENSDART00000099748
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr1_-_52167509 | 1.29 |
ENSDART00000184935
|
BX901942.2
|
|
| chr5_+_60928576 | 1.28 |
ENSDART00000131041
|
doc2b
|
double C2-like domains, beta |
| chr2_-_44971551 | 1.26 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr1_-_46875493 | 1.25 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr24_-_8214253 | 1.17 |
ENSDART00000160432
|
CABZ01077956.1
|
|
| chr6_-_29515709 | 1.16 |
ENSDART00000180205
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr1_+_54115839 | 1.15 |
ENSDART00000180214
|
LO017722.2
|
|
| chr5_+_24156170 | 1.13 |
ENSDART00000136570
|
slc25a15b
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15b |
| chr2_+_16585549 | 1.12 |
ENSDART00000134135
|
si:dkeyp-13a3.10
|
si:dkeyp-13a3.10 |
| chr23_-_45504991 | 1.10 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr16_-_11859309 | 1.09 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr11_+_12175162 | 1.07 |
ENSDART00000125446
|
si:ch211-156l18.7
|
si:ch211-156l18.7 |
| chr1_+_19332837 | 1.06 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr2_-_38282079 | 1.05 |
ENSDART00000145808
|
rnf212b
|
si:ch211-10e2.1 |
| chr18_-_42333428 | 1.02 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr9_+_35860975 | 1.02 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr23_-_28141419 | 1.01 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr2_-_41562868 | 1.01 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr4_-_2975461 | 1.00 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr23_+_24272421 | 0.98 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr14_+_35023923 | 0.97 |
ENSDART00000172171
|
ebf3a
|
early B cell factor 3a |
| chr14_+_30413312 | 0.96 |
ENSDART00000186864
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr1_-_23110740 | 0.95 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr2_-_56649883 | 0.92 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr5_-_13076779 | 0.92 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr8_+_8699085 | 0.88 |
ENSDART00000021209
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr8_+_20825987 | 0.87 |
ENSDART00000133309
|
si:ch211-133l5.4
|
si:ch211-133l5.4 |
| chr20_-_1151265 | 0.86 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr3_-_37351225 | 0.84 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr1_+_44826367 | 0.83 |
ENSDART00000146962
|
STX3
|
zgc:165520 |
| chr14_+_36628131 | 0.83 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr4_-_57416684 | 0.81 |
ENSDART00000169434
|
si:dkey-122c11.7
|
si:dkey-122c11.7 |
| chr10_+_11265387 | 0.77 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr1_+_20593653 | 0.72 |
ENSDART00000132440
|
si:ch211-142c4.1
|
si:ch211-142c4.1 |
| chr3_-_54500354 | 0.71 |
ENSDART00000124215
|
trip10a
|
thyroid hormone receptor interactor 10a |
| chr24_-_6628359 | 0.70 |
ENSDART00000169731
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr5_-_67721368 | 0.70 |
ENSDART00000134386
|
arhgap31
|
Rho GTPase activating protein 31 |
| chr10_-_29892486 | 0.69 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr18_-_4957022 | 0.68 |
ENSDART00000101686
|
zbbx
|
zinc finger, B-box domain containing |
| chr5_-_10007897 | 0.68 |
ENSDART00000109052
|
CR936408.1
|
Danio rerio uncharacterized LOC799523 (LOC799523), mRNA. |
| chr7_+_42206847 | 0.66 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr6_+_40775800 | 0.62 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr15_+_33691455 | 0.57 |
ENSDART00000170329
|
hdac12
|
histone deacetylase 12 |
| chr10_+_36354863 | 0.52 |
ENSDART00000159675
|
or104-1
|
odorant receptor, family C, subfamily 104, member 1 |
| chr23_+_36063599 | 0.52 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr23_+_27068225 | 0.51 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr19_+_19652439 | 0.51 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr7_+_20524064 | 0.50 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr7_-_24005268 | 0.50 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr21_+_37411547 | 0.50 |
ENSDART00000076320
|
mrps17
|
mitochondrial ribosomal protein S17 |
| chr16_-_16619854 | 0.48 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr14_+_33882973 | 0.47 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr10_-_20524592 | 0.45 |
ENSDART00000185048
|
ddhd2
|
DDHD domain containing 2 |
| chr7_-_24046999 | 0.44 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr12_+_30368145 | 0.44 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr19_+_14059349 | 0.41 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr10_+_7563755 | 0.40 |
ENSDART00000165877
|
purg
|
purine-rich element binding protein G |
| chr7_-_31794476 | 0.37 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr5_+_59494079 | 0.31 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr24_-_1021318 | 0.29 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr8_+_53120278 | 0.29 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_-_21287724 | 0.29 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr17_+_30546579 | 0.29 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr9_-_3653259 | 0.23 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr18_-_2433011 | 0.23 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr8_-_23578660 | 0.21 |
ENSDART00000039080
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr8_-_42238543 | 0.20 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr4_-_8060962 | 0.15 |
ENSDART00000146622
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr5_+_29652198 | 0.15 |
ENSDART00000184083
|
tsc1a
|
TSC complex subunit 1a |
| chr7_+_22702225 | 0.13 |
ENSDART00000173672
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr10_-_20524387 | 0.13 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr21_+_4320769 | 0.13 |
ENSDART00000168553
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr24_-_6024466 | 0.10 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr1_-_40102836 | 0.09 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr1_-_53988017 | 0.09 |
ENSDART00000003097
|
RHOU (1 of many)
|
si:ch211-133l11.10 |
| chr19_-_703898 | 0.06 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr6_+_3282809 | 0.02 |
ENSDART00000187444
ENSDART00000187407 ENSDART00000191883 |
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr5_+_61738276 | 0.01 |
ENSDART00000186256
|
RASL10B
|
RAS like family 10 member B |
| chr20_-_45722895 | 0.00 |
ENSDART00000153273
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 13.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 1.9 | 20.9 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 1.4 | 5.8 | GO:0051193 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 1.3 | 6.6 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 1.1 | 5.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.9 | 10.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.7 | 5.5 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.6 | 17.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.5 | 1.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.4 | 1.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.4 | 2.4 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.4 | 2.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.4 | 7.7 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.4 | 1.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.4 | 1.9 | GO:2000379 | protein import into mitochondrial inner membrane(GO:0045039) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.4 | 3.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.3 | 2.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.3 | 4.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.3 | 1.6 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 7.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 5.8 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.3 | 1.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 7.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.2 | 1.7 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 2.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 3.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.2 | 0.9 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.2 | 1.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 1.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 1.4 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 1.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 2.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 3.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 1.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 2.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 7.4 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 6.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.7 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 4.3 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 3.8 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 3.1 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 1.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 1.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 1.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.5 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 10.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 1.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 2.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 1.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 1.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 3.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.0 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 3.1 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.3 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 5.0 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 1.0 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 2.4 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 2.5 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 2.8 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 6.0 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 1.7 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.0 | 3.8 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.7 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 20.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.8 | 9.1 | GO:0043034 | costamere(GO:0043034) |
| 0.7 | 4.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.6 | 11.3 | GO:0031430 | M band(GO:0031430) |
| 0.6 | 14.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.5 | 3.5 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.4 | 2.4 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 7.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 5.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 19.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 3.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 10.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 2.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 7.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 1.9 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 7.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 3.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.7 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 10.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 2.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 0.8 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 3.6 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 5.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 4.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.5 | GO:0005911 | cell-cell junction(GO:0005911) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 20.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.3 | 6.6 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 1.3 | 10.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.3 | 13.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.9 | 3.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.8 | 3.3 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.5 | 1.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.5 | 1.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.5 | 10.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 3.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.4 | 5.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 3.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.3 | 5.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.6 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.2 | 0.9 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 2.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 3.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 1.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 1.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 7.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.5 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 1.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 4.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 2.5 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.7 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 2.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 3.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.0 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 5.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 15.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 30.3 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 2.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 4.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 3.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 4.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 6.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 2.6 | GO:0008092 | cytoskeletal protein binding(GO:0008092) |
| 0.0 | 2.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.0 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 11.9 | GO:0005102 | receptor binding(GO:0005102) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 4.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 10.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.2 | 5.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 2.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 3.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 2.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |