Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
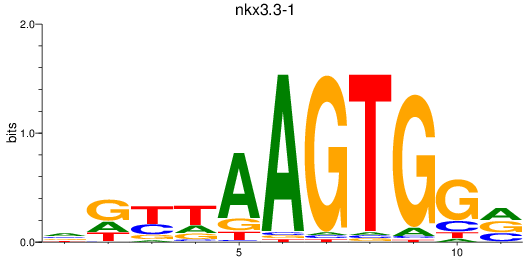
Results for nkx3.3-1
Z-value: 1.18
Transcription factors associated with nkx3.3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3.3-1
|
ENSDARG00000110589 | NK3 homeobox 3 |
Activity profile of nkx3.3-1 motif
Sorted Z-values of nkx3.3-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_2815021 | 15.36 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr7_-_23777445 | 12.30 |
ENSDART00000173527
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr2_+_51028269 | 11.94 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr12_+_31713239 | 11.68 |
ENSDART00000122379
|
habp2
|
hyaluronan binding protein 2 |
| chr8_-_14554785 | 10.79 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr22_+_16022211 | 10.66 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr5_+_37978501 | 10.59 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr8_-_50147948 | 10.29 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr7_+_39446247 | 9.85 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr4_-_8030583 | 8.85 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr7_+_7048245 | 8.76 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr15_+_45563491 | 8.62 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr11_+_42556395 | 8.17 |
ENSDART00000039206
|
rps23
|
ribosomal protein S23 |
| chr10_+_25726694 | 7.74 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr4_-_14642379 | 7.72 |
ENSDART00000114977
|
si:ch211-127b11.1
|
si:ch211-127b11.1 |
| chr22_-_26236188 | 7.64 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr25_+_10547228 | 7.36 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr9_-_7390388 | 7.27 |
ENSDART00000132392
|
slc23a3
|
solute carrier family 23, member 3 |
| chr25_+_28282274 | 7.25 |
ENSDART00000164502
|
aass
|
aminoadipate-semialdehyde synthase |
| chr19_+_30990129 | 7.10 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr3_+_29640837 | 7.09 |
ENSDART00000132298
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr15_-_41245962 | 6.88 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr2_+_30948503 | 6.78 |
ENSDART00000149923
|
myom1a
|
myomesin 1a (skelemin) |
| chr19_+_30990815 | 6.52 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr5_+_13870340 | 6.35 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr14_+_17376940 | 6.02 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr12_-_3053699 | 5.95 |
ENSDART00000139721
|
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr13_-_2215213 | 5.94 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr14_+_26719691 | 5.91 |
ENSDART00000078522
ENSDART00000172927 |
eef1g
|
eukaryotic translation elongation factor 1 gamma |
| chr4_-_14649158 | 5.89 |
ENSDART00000145737
|
si:dkey-183c2.4
|
si:dkey-183c2.4 |
| chr24_-_26484298 | 5.70 |
ENSDART00000140647
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr15_-_3277635 | 5.59 |
ENSDART00000189094
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr17_+_6563307 | 5.55 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr20_+_23498255 | 5.50 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr16_+_16977786 | 5.44 |
ENSDART00000043173
ENSDART00000132150 |
rpl18
|
ribosomal protein L18 |
| chr25_+_11456696 | 5.44 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr5_-_12063381 | 5.44 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr10_+_10387328 | 5.43 |
ENSDART00000080904
|
sardh
|
sarcosine dehydrogenase |
| chr25_-_37284370 | 5.28 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr23_+_25292147 | 5.24 |
ENSDART00000131486
|
pa2g4b
|
proliferation-associated 2G4, b |
| chr21_-_28523548 | 5.23 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr15_+_46606090 | 5.18 |
ENSDART00000020921
|
CABZ01044048.1
|
|
| chr18_+_24562188 | 5.03 |
ENSDART00000099463
|
lysmd4
|
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr9_-_43073960 | 5.02 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr21_+_26612777 | 4.96 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr24_-_33308045 | 4.88 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr19_+_43604643 | 4.86 |
ENSDART00000151168
|
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr15_+_17756113 | 4.82 |
ENSDART00000155197
|
si:ch211-213d14.2
|
si:ch211-213d14.2 |
| chr22_+_38194151 | 4.75 |
ENSDART00000121965
|
cp
|
ceruloplasmin |
| chr15_+_5973909 | 4.66 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr19_+_791538 | 4.62 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr9_-_49531762 | 4.61 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr22_+_32228882 | 4.60 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr1_-_54706039 | 4.49 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr16_-_38609146 | 4.48 |
ENSDART00000144651
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr16_+_3982590 | 4.40 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr19_-_34970312 | 4.39 |
ENSDART00000102896
|
ndrg1a
|
N-myc downstream regulated 1a |
| chr18_+_29156827 | 4.33 |
ENSDART00000137587
ENSDART00000135633 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr5_+_44846280 | 4.23 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr22_+_7480465 | 4.23 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr25_+_10416583 | 4.12 |
ENSDART00000073907
|
ehf
|
ets homologous factor |
| chr22_-_36770649 | 4.10 |
ENSDART00000169442
|
acy1
|
aminoacylase 1 |
| chr9_+_15893093 | 4.08 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr25_+_20091021 | 4.07 |
ENSDART00000187545
ENSDART00000053265 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr4_-_75982054 | 4.01 |
ENSDART00000168369
|
si:ch211-232d10.1
|
si:ch211-232d10.1 |
| chr25_-_16600811 | 3.87 |
ENSDART00000011397
|
cpa2
|
carboxypeptidase A2 (pancreatic) |
| chr16_+_46725087 | 3.87 |
ENSDART00000008920
|
rab11al
|
RAB11a, member RAS oncogene family, like |
| chr12_+_19030391 | 3.82 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr4_+_16715267 | 3.78 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr10_+_10386435 | 3.76 |
ENSDART00000179214
ENSDART00000189799 ENSDART00000193875 |
sardh
|
sarcosine dehydrogenase |
| chr22_-_20376488 | 3.74 |
ENSDART00000140187
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr2_+_39618951 | 3.62 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr9_-_55586151 | 3.62 |
ENSDART00000181886
|
arsh
|
arylsulfatase H |
| chr10_+_40660772 | 3.51 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr18_+_19842569 | 3.50 |
ENSDART00000147470
|
iqch
|
IQ motif containing H |
| chr16_-_13612650 | 3.48 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr21_+_5993188 | 3.46 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr6_-_43028270 | 3.37 |
ENSDART00000149092
ENSDART00000149559 |
glyctk
|
glycerate kinase |
| chr21_+_21812311 | 3.37 |
ENSDART00000151253
|
neu3.4
|
sialidase 3 (membrane sialidase), tandem duplicate 4 |
| chr20_+_826459 | 3.34 |
ENSDART00000104740
|
nt5e
|
5'-nucleotidase, ecto (CD73) |
| chr3_-_32912787 | 3.33 |
ENSDART00000156149
|
g6pcb
|
glucose-6-phosphatase b, catalytic subunit |
| chr5_-_57204352 | 3.31 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr24_-_25166720 | 3.30 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr12_+_22580579 | 3.28 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr25_-_19666107 | 3.27 |
ENSDART00000149889
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr19_-_6840506 | 3.25 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr1_-_58887610 | 3.22 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr13_-_280827 | 3.16 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr16_-_26132122 | 3.10 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr20_-_7992496 | 3.06 |
ENSDART00000145521
|
plpp3
|
phospholipid phosphatase 3 |
| chr18_+_33675664 | 3.05 |
ENSDART00000140043
|
si:dkey-47k20.8
|
si:dkey-47k20.8 |
| chr7_-_59311165 | 3.01 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr9_-_53666031 | 3.00 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr17_+_15388479 | 3.00 |
ENSDART00000052439
|
si:ch211-266g18.6
|
si:ch211-266g18.6 |
| chr24_+_15670013 | 2.99 |
ENSDART00000185826
|
CU929414.1
|
|
| chr5_-_23855447 | 2.98 |
ENSDART00000051541
|
gbgt1l3
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 3 |
| chr4_-_10835620 | 2.97 |
ENSDART00000150739
|
ppfibp1a
|
PTPRF interacting protein, binding protein 1a (liprin beta 1) |
| chr16_+_27444098 | 2.95 |
ENSDART00000157690
|
invs
|
inversin |
| chr1_+_35862550 | 2.93 |
ENSDART00000132118
|
si:ch211-194g2.4
|
si:ch211-194g2.4 |
| chr9_-_34882516 | 2.93 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr16_-_9675982 | 2.87 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
| chr16_+_38240027 | 2.83 |
ENSDART00000111081
|
prune
|
prune exopolyphosphatase |
| chr4_-_12862087 | 2.83 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr9_+_8380728 | 2.82 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr13_-_40237121 | 2.82 |
ENSDART00000145635
|
loxl4
|
lysyl oxidase-like 4 |
| chr10_+_40737540 | 2.75 |
ENSDART00000125577
|
taar19a
|
trace amine associated receptor 19a |
| chr21_-_41873065 | 2.74 |
ENSDART00000014538
|
endou2
|
endonuclease, polyU-specific 2 |
| chr5_+_58550291 | 2.72 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr2_-_55797318 | 2.72 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr25_+_2361721 | 2.70 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr5_-_8907819 | 2.65 |
ENSDART00000188523
|
adamts12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr16_-_22585289 | 2.65 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr6_+_3280939 | 2.64 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr21_+_5589923 | 2.63 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr10_+_40711286 | 2.62 |
ENSDART00000140242
|
si:ch211-139n6.11
|
si:ch211-139n6.11 |
| chr4_-_11098539 | 2.60 |
ENSDART00000135511
|
si:dkey-21h14.12
|
si:dkey-21h14.12 |
| chr10_-_39052264 | 2.57 |
ENSDART00000144036
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr19_+_7810028 | 2.56 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr8_+_53051701 | 2.55 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr14_-_36763302 | 2.53 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr23_-_46040618 | 2.52 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr4_+_9467049 | 2.45 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr4_-_71708567 | 2.38 |
ENSDART00000182645
|
si:dkeyp-4f2.1
|
si:dkeyp-4f2.1 |
| chr7_-_27038488 | 2.38 |
ENSDART00000052731
ENSDART00000191382 |
nucb2a
|
nucleobindin 2a |
| chr18_-_33093705 | 2.38 |
ENSDART00000059500
|
olfcd3
|
olfactory receptor C family, d3 |
| chr2_-_57900430 | 2.38 |
ENSDART00000132245
ENSDART00000140060 |
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
| chr12_+_17754859 | 2.38 |
ENSDART00000112119
|
bhlha15
|
basic helix-loop-helix family, member a15 |
| chr24_-_41195068 | 2.36 |
ENSDART00000121592
|
acvr2ba
|
activin A receptor type 2Ba |
| chr2_+_53114476 | 2.35 |
ENSDART00000080850
ENSDART00000104187 |
CR384061.1
|
|
| chr22_+_6323600 | 2.35 |
ENSDART00000193061
|
AL928685.4
|
|
| chr17_+_23554932 | 2.31 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr25_-_12635371 | 2.31 |
ENSDART00000162463
|
zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr5_-_54712159 | 2.28 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr23_-_17003533 | 2.27 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr15_+_15771418 | 2.24 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr21_+_30549512 | 2.23 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr1_+_56779495 | 2.22 |
ENSDART00000192871
|
FO680692.1
|
|
| chr21_+_21702623 | 2.21 |
ENSDART00000136840
|
or125-2
|
odorant receptor, family E, subfamily 125, member 2 |
| chr17_+_48164536 | 2.20 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr7_+_17393794 | 2.20 |
ENSDART00000059673
|
nitr7b
|
novel immune-type receptor 7b |
| chr2_-_51454868 | 2.16 |
ENSDART00000172550
|
si:dkeyp-104b3.2
|
si:dkeyp-104b3.2 |
| chr11_+_31680513 | 2.15 |
ENSDART00000139900
ENSDART00000040305 |
diaph3
|
diaphanous-related formin 3 |
| chr1_+_55643198 | 2.15 |
ENSDART00000060693
|
adgre7
|
adhesion G protein-coupled receptor E7 |
| chr23_+_7518294 | 2.15 |
ENSDART00000081536
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr23_-_26227805 | 2.14 |
ENSDART00000158082
|
BX927204.1
|
|
| chr21_-_43094210 | 2.12 |
ENSDART00000144747
|
si:ch73-68b22.2
|
si:ch73-68b22.2 |
| chr12_-_16990896 | 2.11 |
ENSDART00000152402
|
ifit12
|
interferon-induced protein with tetratricopeptide repeats 12 |
| chr10_-_33343244 | 2.10 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr21_+_25344374 | 2.10 |
ENSDART00000144724
|
or132-2
|
odorant receptor, family H, subfamily 132, member 2 |
| chr3_-_34066548 | 2.08 |
ENSDART00000151302
|
ighv9-2
|
immunoglobulin heavy variable 9-2 |
| chr18_+_33100606 | 2.08 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr18_-_28938912 | 2.06 |
ENSDART00000136201
|
si:ch211-174j14.2
|
si:ch211-174j14.2 |
| chr1_-_10847753 | 2.06 |
ENSDART00000141052
|
dmd
|
dystrophin |
| chr15_+_37972671 | 2.04 |
ENSDART00000166764
|
si:dkey-238d18.9
|
si:dkey-238d18.9 |
| chr4_+_53146060 | 2.04 |
ENSDART00000124912
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr13_+_33462232 | 2.04 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr5_-_24124118 | 2.03 |
ENSDART00000051550
|
capga
|
capping protein (actin filament), gelsolin-like a |
| chr21_-_40040178 | 2.02 |
ENSDART00000159741
|
BX248108.1
|
Danio rerio multidrug and toxin extrusion protein 1-like (mate4), mRNA. |
| chr5_+_37890521 | 2.01 |
ENSDART00000140207
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr22_+_38159823 | 2.01 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr6_+_58622831 | 2.01 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr19_+_42470396 | 1.95 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr22_-_31020690 | 1.93 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr3_+_11926030 | 1.93 |
ENSDART00000081367
|
dnaja3a
|
DnaJ (Hsp40) homolog, subfamily A, member 3A |
| chr3_-_61592417 | 1.93 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr18_-_31423569 | 1.93 |
ENSDART00000185028
|
CU570893.1
|
|
| chr4_+_53145525 | 1.93 |
ENSDART00000166960
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr25_+_28772632 | 1.92 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr8_+_31435452 | 1.92 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr22_+_19522982 | 1.88 |
ENSDART00000192428
ENSDART00000190812 |
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr18_+_2593756 | 1.87 |
ENSDART00000158022
|
p2ry2.3
|
purinergic receptor P2Y2, tandem duplicate 3 |
| chr5_-_23783739 | 1.86 |
ENSDART00000139502
|
GBGT1 (1 of many)
|
si:ch211-287c22.1 |
| chr3_-_5393909 | 1.86 |
ENSDART00000031885
|
si:ch73-106l15.2
|
si:ch73-106l15.2 |
| chr4_+_13931578 | 1.85 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr7_-_70283372 | 1.84 |
ENSDART00000112969
|
adgra3
|
adhesion G protein-coupled receptor A3 |
| chr4_+_71382288 | 1.84 |
ENSDART00000181926
|
si:ch211-76m11.8
|
si:ch211-76m11.8 |
| chr10_+_35358675 | 1.83 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr17_-_2811992 | 1.82 |
ENSDART00000067543
|
gpr65
|
G protein-coupled receptor 65 |
| chr8_+_23788981 | 1.82 |
ENSDART00000144229
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr16_-_12097558 | 1.77 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr5_-_69425224 | 1.76 |
ENSDART00000158096
|
FQ311924.1
|
|
| chr10_-_40964739 | 1.71 |
ENSDART00000076359
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr22_-_16755885 | 1.64 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr7_+_12835048 | 1.63 |
ENSDART00000016465
|
cx36.7
|
connexin 36.7 |
| chr2_+_24931677 | 1.62 |
ENSDART00000021528
|
agtr1a
|
angiotensin II receptor, type 1a |
| chr3_-_2091029 | 1.60 |
ENSDART00000141464
|
si:dkey-88j15.4
|
si:dkey-88j15.4 |
| chr1_+_524388 | 1.60 |
ENSDART00000020327
|
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr10_+_40774215 | 1.58 |
ENSDART00000131493
|
taar19b
|
trace amine associated receptor 19b |
| chr20_-_33675676 | 1.58 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr12_-_44016898 | 1.58 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr15_+_33991928 | 1.57 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr13_-_50463938 | 1.57 |
ENSDART00000083857
|
ccnj
|
cyclin J |
| chr12_+_33433882 | 1.56 |
ENSDART00000185053
|
fasn
|
fatty acid synthase |
| chr4_-_62503772 | 1.55 |
ENSDART00000108891
|
si:dkey-165b20.1
|
si:dkey-165b20.1 |
| chr2_+_45593783 | 1.52 |
ENSDART00000148037
|
fndc7rs2
|
fibronectin type III domain containing 7, related sequence 2 |
| chr25_-_11016675 | 1.52 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr1_-_11075403 | 1.51 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr4_-_49875682 | 1.51 |
ENSDART00000185921
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr22_+_19188809 | 1.50 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr7_-_17267950 | 1.49 |
ENSDART00000122171
|
nitr10a
|
novel immune-type receptor 10a |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3.3-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 3.1 | 9.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 2.6 | 15.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 2.4 | 7.2 | GO:0033512 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 2.4 | 7.1 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 2.1 | 10.6 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 2.0 | 5.9 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 1.9 | 5.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 1.1 | 4.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.8 | 5.7 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.7 | 2.9 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.6 | 4.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.6 | 3.0 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.6 | 5.3 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.6 | 2.9 | GO:0045056 | transcytosis(GO:0045056) |
| 0.6 | 3.3 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.6 | 3.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.5 | 3.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.5 | 1.9 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.5 | 2.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.4 | 9.7 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.4 | 1.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.4 | 4.7 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.4 | 12.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.4 | 2.0 | GO:0097094 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.4 | 9.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.4 | 1.9 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.4 | 2.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 8.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 8.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.3 | 1.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 2.6 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 5.0 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.3 | 17.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.3 | 3.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.3 | 4.9 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.3 | 2.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 2.0 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 2.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 2.4 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.2 | 1.8 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.2 | 3.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 1.8 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.2 | 4.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 4.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 3.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 3.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.8 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.2 | 3.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.2 | 5.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.2 | 1.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 2.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.6 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 11.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 1.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 4.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 5.0 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.1 | 2.1 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 3.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 15.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 0.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.5 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 4.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 3.8 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 1.6 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.1 | 0.3 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.2 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 1.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 4.2 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 7.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 3.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.9 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 3.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.8 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 3.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 2.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 3.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 4.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.4 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 1.0 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.9 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 1.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 12.9 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.9 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 4.1 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 2.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 4.1 | GO:0006520 | cellular amino acid metabolic process(GO:0006520) |
| 0.0 | 1.3 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 1.0 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 2.3 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 0.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 1.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 1.1 | GO:1903706 | regulation of hemopoiesis(GO:1903706) |
| 0.0 | 9.0 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 2.1 | GO:0045087 | innate immune response(GO:0045087) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 11.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.1 | 12.3 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.9 | 4.5 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.8 | 10.6 | GO:0042627 | chylomicron(GO:0042627) |
| 0.8 | 2.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.4 | 7.1 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.4 | 4.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 13.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 6.8 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 3.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 4.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 3.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 5.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 2.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 8.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 3.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 5.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 11.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 3.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.7 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 11.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 6.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 4.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 4.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 8.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 5.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 3.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 11.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 5.0 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 3.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 5.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 3.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 8.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 45.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 4.4 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 12.7 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.9 | GO:0044441 | ciliary part(GO:0044441) |
| 0.0 | 1.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 1.6 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 3.6 | 10.8 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 2.4 | 7.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 2.3 | 9.2 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 2.1 | 10.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.5 | 5.9 | GO:0005521 | lamin binding(GO:0005521) |
| 1.1 | 6.4 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.9 | 12.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.9 | 9.9 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.8 | 3.4 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.6 | 3.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.6 | 5.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.5 | 2.6 | GO:2001070 | starch binding(GO:2001070) |
| 0.5 | 23.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.5 | 2.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.5 | 5.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.5 | 1.4 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.4 | 1.6 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.4 | 2.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.4 | 2.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 1.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 8.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 2.6 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.3 | 4.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 3.4 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.3 | 7.2 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.2 | 1.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 1.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 3.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 3.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 5.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.2 | 1.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 1.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 2.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 2.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 5.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 2.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 3.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 2.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 3.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 3.3 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.1 | 10.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 3.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.6 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 7.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 3.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 5.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 7.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 3.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 3.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 24.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 18.3 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 3.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 14.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 6.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 3.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 4.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.4 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 6.7 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.8 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 10.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 8.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 2.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 1.3 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 3.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.3 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 8.7 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 2.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.5 | GO:0005507 | copper ion binding(GO:0005507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 11.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.3 | 14.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 18.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 10.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.7 | 3.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 4.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 9.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 15.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 4.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 2.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 7.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 4.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 3.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 5.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 11.4 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 2.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 2.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 6.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |