Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
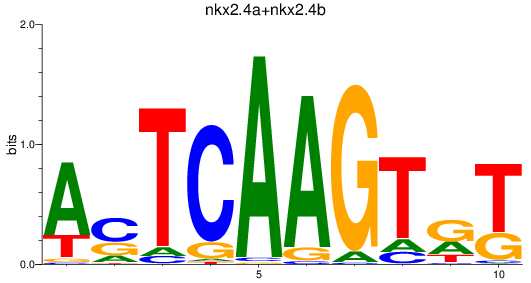
Results for nkx2.4a+nkx2.4b
Z-value: 0.46
Transcription factors associated with nkx2.4a+nkx2.4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.4a
|
ENSDARG00000075107 | NK2 homeobox 4a |
|
nkx2.4b
|
ENSDARG00000104107 | NK2 homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.4a | dr11_v1_chr17_-_42097267_42097267 | 0.13 | 2.2e-01 | Click! |
| nkx2.4b | dr11_v1_chr20_+_48803248_48803248 | 0.11 | 2.9e-01 | Click! |
Activity profile of nkx2.4a+nkx2.4b motif
Sorted Z-values of nkx2.4a+nkx2.4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_22139566 | 3.11 |
ENSDART00000149017
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr1_-_38816685 | 2.74 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr7_-_35708450 | 2.63 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr13_-_51846224 | 2.43 |
ENSDART00000184663
|
LT631684.2
|
|
| chr5_-_28915130 | 2.39 |
ENSDART00000078592
|
npdc1b
|
neural proliferation, differentiation and control, 1b |
| chr1_-_14234076 | 1.92 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr17_+_27456804 | 1.88 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr23_-_35347714 | 1.71 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr8_-_30316436 | 1.59 |
ENSDART00000111358
ENSDART00000167065 |
zgc:162939
|
zgc:162939 |
| chr2_-_898899 | 1.56 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr12_-_20362041 | 1.55 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr24_-_28259127 | 1.49 |
ENSDART00000149589
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr9_-_23033818 | 1.46 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr17_-_31044803 | 1.43 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr12_+_27243059 | 1.36 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr1_-_14233815 | 1.34 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr24_+_19210001 | 1.33 |
ENSDART00000179373
ENSDART00000139299 |
zgc:162928
|
zgc:162928 |
| chr11_+_24046179 | 1.32 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr5_+_34981584 | 1.32 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr19_-_27776649 | 1.22 |
ENSDART00000135348
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr21_+_27189490 | 1.20 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr1_-_38171648 | 1.11 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr7_+_25323742 | 1.11 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr8_-_49345388 | 1.08 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr10_+_21807497 | 1.05 |
ENSDART00000164634
|
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr5_-_33460959 | 1.04 |
ENSDART00000085636
|
si:ch211-182d3.1
|
si:ch211-182d3.1 |
| chr19_+_23982466 | 0.98 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr4_+_22365061 | 0.97 |
ENSDART00000039277
|
lhfpl3
|
LHFPL tetraspan subfamily member 3 |
| chr17_+_8184649 | 0.97 |
ENSDART00000091818
|
tulp4b
|
tubby like protein 4b |
| chr12_+_5977777 | 0.96 |
ENSDART00000152302
|
si:ch211-131k2.2
|
si:ch211-131k2.2 |
| chr22_+_39096911 | 0.93 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr10_-_10863936 | 0.91 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr4_+_65649267 | 0.89 |
ENSDART00000184068
|
si:dkey-205i10.1
|
si:dkey-205i10.1 |
| chr7_-_35126374 | 0.88 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr6_+_374875 | 0.87 |
ENSDART00000171698
|
si:zfos-169g10.3
|
si:zfos-169g10.3 |
| chr10_+_5268054 | 0.87 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr17_+_43032529 | 0.85 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr10_-_10864331 | 0.84 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr12_-_28848015 | 0.84 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr6_-_13114821 | 0.82 |
ENSDART00000164579
ENSDART00000165375 |
zgc:194469
|
zgc:194469 |
| chr12_+_49100365 | 0.82 |
ENSDART00000171905
|
CABZ01075125.1
|
|
| chr8_+_23485079 | 0.81 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr14_+_23158021 | 0.81 |
ENSDART00000084664
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr22_+_8753092 | 0.80 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr16_-_11766265 | 0.77 |
ENSDART00000141547
|
zgc:158701
|
zgc:158701 |
| chr8_-_17516448 | 0.74 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr8_+_36803415 | 0.73 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr11_-_30352333 | 0.72 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr23_+_6795709 | 0.71 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr22_+_3223489 | 0.71 |
ENSDART00000082011
|
lim2.2
|
lens intrinsic membrane protein 2.2 |
| chr14_+_14225048 | 0.70 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr7_+_60551133 | 0.70 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr23_+_39346930 | 0.70 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr8_+_39607466 | 0.69 |
ENSDART00000097427
|
msi1
|
musashi RNA-binding protein 1 |
| chr15_-_47937204 | 0.68 |
ENSDART00000154705
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr22_-_7457247 | 0.67 |
ENSDART00000106081
|
BX511034.2
|
|
| chr19_-_25113660 | 0.65 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr14_-_8816933 | 0.63 |
ENSDART00000163383
|
si:dkeyp-115e12.7
|
si:dkeyp-115e12.7 |
| chr11_+_39120306 | 0.61 |
ENSDART00000155746
ENSDART00000154158 |
cdc42
|
cell division cycle 42 |
| chr7_+_39416336 | 0.61 |
ENSDART00000171783
|
CT030188.1
|
|
| chr14_+_23709134 | 0.61 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr25_+_19485198 | 0.60 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr15_-_12011390 | 0.59 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr14_+_23709543 | 0.58 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr1_+_45671687 | 0.58 |
ENSDART00000146101
|
mcoln1a
|
mucolipin 1a |
| chr10_+_8481660 | 0.57 |
ENSDART00000109559
|
BX682234.1
|
|
| chr5_+_53824959 | 0.57 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr10_-_15048781 | 0.56 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr21_+_30084823 | 0.55 |
ENSDART00000154573
|
prob1
|
proline-rich basic protein 1 |
| chr22_-_28373698 | 0.55 |
ENSDART00000157592
|
si:ch211-213c4.5
|
si:ch211-213c4.5 |
| chr12_-_11768889 | 0.54 |
ENSDART00000057866
|
atoh1c
|
atonal bHLH transcription factor 1c |
| chr2_+_24203229 | 0.54 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr15_+_17441734 | 0.54 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr6_-_42073367 | 0.53 |
ENSDART00000154304
|
grm2a
|
glutamate receptor, metabotropic 2a |
| chr4_-_60312069 | 0.52 |
ENSDART00000167207
|
si:dkey-248e17.4
|
si:dkey-248e17.4 |
| chr7_-_26049282 | 0.52 |
ENSDART00000136389
ENSDART00000101124 |
rnaseka
|
ribonuclease, RNase K a |
| chr3_+_33340939 | 0.52 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr17_-_42213285 | 0.52 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr20_+_27020201 | 0.51 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr10_+_9346383 | 0.51 |
ENSDART00000064975
ENSDART00000141869 |
rbm18
|
RNA binding motif protein 18 |
| chr6_+_58832155 | 0.51 |
ENSDART00000144842
|
dctn2
|
dynactin 2 (p50) |
| chr17_-_7496341 | 0.50 |
ENSDART00000186907
|
si:ch211-278p9.3
|
si:ch211-278p9.3 |
| chr13_-_2112008 | 0.48 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr8_-_22558773 | 0.48 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr14_-_30876708 | 0.45 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr7_-_5039562 | 0.45 |
ENSDART00000154438
|
ltb4r
|
leukotriene B4 receptor |
| chr16_+_11151699 | 0.45 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr9_+_18716485 | 0.44 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr7_+_26049818 | 0.42 |
ENSDART00000173611
|
si:dkey-6n21.12
|
si:dkey-6n21.12 |
| chr5_-_54554583 | 0.42 |
ENSDART00000158865
ENSDART00000158069 |
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr21_+_17110598 | 0.40 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr11_+_36158134 | 0.40 |
ENSDART00000189827
ENSDART00000163330 |
grm2b
|
glutamate receptor, metabotropic 2b |
| chr23_-_18982499 | 0.39 |
ENSDART00000012507
|
bcl2l1
|
bcl2-like 1 |
| chr15_+_850829 | 0.39 |
ENSDART00000157374
|
si:dkey-77f5.11
|
si:dkey-77f5.11 |
| chr15_-_23485752 | 0.38 |
ENSDART00000152706
|
nlrx1
|
NLR family member X1 |
| chr13_-_48388726 | 0.37 |
ENSDART00000169473
|
fbxo11a
|
F-box protein 11a |
| chr1_+_22654875 | 0.37 |
ENSDART00000019698
ENSDART00000161874 |
anxa5b
|
annexin A5b |
| chr2_-_22575851 | 0.37 |
ENSDART00000182932
|
FQ377628.2
|
|
| chr8_-_39859688 | 0.36 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr8_+_53120278 | 0.36 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_-_59232267 | 0.35 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr3_+_24134418 | 0.34 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr14_+_8343498 | 0.34 |
ENSDART00000164551
|
nrg2b
|
neuregulin 2b |
| chr15_+_22722684 | 0.34 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr23_+_39346774 | 0.34 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr19_-_3261019 | 0.34 |
ENSDART00000134407
|
si:ch211-133n4.12
|
si:ch211-133n4.12 |
| chr4_+_23126558 | 0.33 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr4_-_43731342 | 0.33 |
ENSDART00000146627
|
si:ch211-226o13.3
|
si:ch211-226o13.3 |
| chr15_-_8517555 | 0.32 |
ENSDART00000140213
|
npas1
|
neuronal PAS domain protein 1 |
| chr25_-_1079417 | 0.31 |
ENSDART00000163134
|
FO907089.1
|
|
| chr6_-_13114406 | 0.31 |
ENSDART00000188015
|
zgc:194469
|
zgc:194469 |
| chr6_-_30210378 | 0.31 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr19_-_17303500 | 0.30 |
ENSDART00000162355
|
sf3a3
|
splicing factor 3a, subunit 3 |
| chr5_-_32882162 | 0.30 |
ENSDART00000085769
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr2_+_23731194 | 0.29 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr14_+_14224730 | 0.29 |
ENSDART00000180112
ENSDART00000184891 ENSDART00000174760 |
nlgn3a
|
neuroligin 3a |
| chr20_+_23742574 | 0.27 |
ENSDART00000050559
|
sh3rf1
|
SH3 domain containing ring finger 1 |
| chr9_+_25832329 | 0.26 |
ENSDART00000130059
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr10_-_17179415 | 0.26 |
ENSDART00000131751
|
depdc5
|
DEP domain containing 5 |
| chr21_-_41147818 | 0.25 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr7_+_39664055 | 0.25 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr5_+_36870737 | 0.25 |
ENSDART00000145182
|
slc8a2a
|
solute carrier family 8 (sodium/calcium exchanger), member 2a |
| chr4_+_50410452 | 0.25 |
ENSDART00000146006
|
si:dkey-156k2.8
|
si:dkey-156k2.8 |
| chr10_-_9346360 | 0.24 |
ENSDART00000039536
|
mrrf
|
mitochondrial ribosome recycling factor |
| chr4_+_23127104 | 0.24 |
ENSDART00000139543
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr18_-_7481036 | 0.24 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr16_+_25259313 | 0.23 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr14_-_9982603 | 0.23 |
ENSDART00000054687
|
il1rapl2
|
interleukin 1 receptor accessory protein-like 2 |
| chr23_-_26532696 | 0.23 |
ENSDART00000124811
ENSDART00000180274 |
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr19_+_7938121 | 0.22 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr2_-_51507540 | 0.22 |
ENSDART00000166605
ENSDART00000161093 |
pigrl2.3
|
polymeric immunoglobulin receptor-like 2.3 |
| chr8_-_25120231 | 0.21 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr1_-_7603734 | 0.21 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr1_+_29862074 | 0.20 |
ENSDART00000133905
ENSDART00000136786 ENSDART00000135252 |
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr5_+_56119975 | 0.20 |
ENSDART00000083137
|
mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr17_-_8673278 | 0.20 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr7_+_42206847 | 0.19 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr17_-_42213822 | 0.19 |
ENSDART00000187904
ENSDART00000180029 |
nkx2.2a
|
NK2 homeobox 2a |
| chr11_-_34628789 | 0.19 |
ENSDART00000192433
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr2_-_6292510 | 0.18 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr16_-_34258931 | 0.18 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr16_+_24971717 | 0.18 |
ENSDART00000156958
|
fpr1
|
formyl peptide receptor 1 |
| chr1_-_31657854 | 0.18 |
ENSDART00000085309
|
dpcd
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr13_+_25761279 | 0.18 |
ENSDART00000177818
ENSDART00000002863 |
neurog3
|
neurogenin 3 |
| chr4_+_44454270 | 0.17 |
ENSDART00000150482
|
si:dkeyp-100h4.4
|
si:dkeyp-100h4.4 |
| chr3_+_58354852 | 0.16 |
ENSDART00000155715
|
vwa3a
|
von Willebrand factor A domain containing 3A |
| chr5_-_39171302 | 0.16 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr20_+_27464721 | 0.16 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr5_+_36611128 | 0.15 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr17_-_8673567 | 0.15 |
ENSDART00000192714
ENSDART00000012546 |
ctbp2a
|
C-terminal binding protein 2a |
| chr2_-_32668668 | 0.15 |
ENSDART00000190450
|
puf60a
|
poly-U binding splicing factor a |
| chr17_-_22552678 | 0.15 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr17_-_28097760 | 0.15 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr20_+_35484070 | 0.14 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr1_-_21599219 | 0.13 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr23_+_7505943 | 0.13 |
ENSDART00000135787
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr1_+_31658011 | 0.12 |
ENSDART00000192203
|
poll
|
polymerase (DNA directed), lambda |
| chr10_+_2742499 | 0.12 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr7_-_18601206 | 0.12 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr23_+_34321237 | 0.12 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr8_-_23776399 | 0.12 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr2_-_36604589 | 0.12 |
ENSDART00000167352
|
tradv41.0
|
T-cell receptor alpha/delta variable 41.0 |
| chr2_+_47754419 | 0.11 |
ENSDART00000188221
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr19_-_23470709 | 0.11 |
ENSDART00000138884
|
pimr80
|
Pim proto-oncogene, serine/threonine kinase, related 80 |
| chr11_-_438294 | 0.10 |
ENSDART00000040812
|
nuf2
|
NUF2, NDC80 kinetochore complex component, homolog |
| chr4_-_965267 | 0.10 |
ENSDART00000093289
|
atp23
|
ATP23 metallopeptidase and ATP synthase assembly factor homolog |
| chr14_-_32486757 | 0.09 |
ENSDART00000148830
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr3_-_11008532 | 0.09 |
ENSDART00000165086
|
CR382337.3
|
|
| chr14_-_17622080 | 0.09 |
ENSDART00000112149
|
si:ch211-159i8.4
|
si:ch211-159i8.4 |
| chr15_-_1590858 | 0.09 |
ENSDART00000081875
|
nnr
|
nanor |
| chr12_+_32199651 | 0.08 |
ENSDART00000153027
|
si:ch211-277e21.1
|
si:ch211-277e21.1 |
| chr3_-_39305291 | 0.08 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr5_+_36896933 | 0.08 |
ENSDART00000151984
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr9_+_20869166 | 0.08 |
ENSDART00000147892
|
wdr3
|
WD repeat domain 3 |
| chr1_+_31657842 | 0.08 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr14_+_16083818 | 0.07 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr12_-_1361517 | 0.07 |
ENSDART00000188297
|
LO018020.2
|
|
| chr18_+_23397010 | 0.06 |
ENSDART00000193765
|
mctp2a
|
multiple C2 domains, transmembrane 2a |
| chr4_-_17838179 | 0.05 |
ENSDART00000146931
|
avpr2l
|
arginine vasopressin receptor 2, like |
| chr15_-_28569786 | 0.05 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr20_-_43079478 | 0.04 |
ENSDART00000060982
|
snx9b
|
sorting nexin 9b |
| chr4_-_65747476 | 0.04 |
ENSDART00000142421
|
si:dkey-28k24.2
|
si:dkey-28k24.2 |
| chr19_-_23452345 | 0.04 |
ENSDART00000136875
|
pimr82
|
Pim proto-oncogene, serine/threonine kinase, related 82 |
| chr3_+_32571929 | 0.04 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr17_+_43659940 | 0.03 |
ENSDART00000145738
ENSDART00000075619 |
adob
|
2-aminoethanethiol (cysteamine) dioxygenase b |
| chr20_+_48782068 | 0.03 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr9_+_159720 | 0.02 |
ENSDART00000166215
|
cldn10l2
|
claudin 10-like 2 |
| chr19_+_26424734 | 0.02 |
ENSDART00000187402
|
npr1a
|
natriuretic peptide receptor 1a |
| chr15_-_12011202 | 0.02 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr2_+_36608387 | 0.02 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr12_-_19279103 | 0.01 |
ENSDART00000186669
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
| chr9_-_35155089 | 0.01 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr4_+_17843717 | 0.01 |
ENSDART00000113507
|
apaf1
|
apoptotic peptidase activating factor 1 |
| chr10_-_9086716 | 0.01 |
ENSDART00000140095
|
arl15b
|
ADP-ribosylation factor-like 15b |
| chr22_+_19111444 | 0.01 |
ENSDART00000109655
|
fgf22
|
fibroblast growth factor 22 |
| chr4_-_17680493 | 0.00 |
ENSDART00000180131
|
BX890572.2
|
|
| chr7_+_20966434 | 0.00 |
ENSDART00000185570
|
efnb3b
|
ephrin-B3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.4a+nkx2.4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.4 | 1.2 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.4 | 1.1 | GO:0035992 | tendon formation(GO:0035992) |
| 0.3 | 1.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 2.6 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 0.7 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 1.0 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 0.5 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.2 | 0.8 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.5 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 1.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 1.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 0.9 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.6 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 1.0 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.4 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.3 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.6 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.3 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.1 | 0.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.5 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 2.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.9 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 1.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.0 | 0.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.6 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.2 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 1.9 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.4 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.5 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.8 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.9 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.2 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.5 | GO:0038127 | epidermal growth factor receptor signaling pathway(GO:0007173) ERBB signaling pathway(GO:0038127) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 1.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 2.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 1.0 | GO:0031834 | neurokinin receptor binding(GO:0031834) substance P receptor binding(GO:0031835) |
| 0.2 | 1.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.9 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.5 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.6 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 3.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 1.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 1.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.9 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 2.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.9 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 2.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.1 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.5 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |