Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
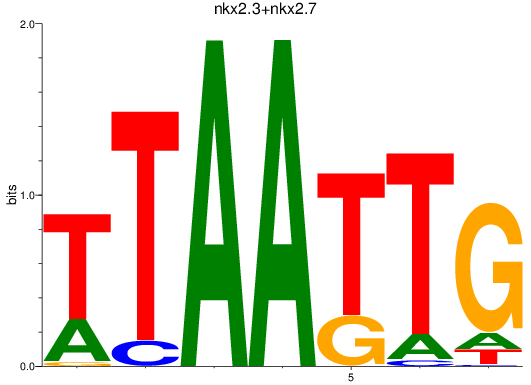
Results for nkx2.3+nkx2.7
Z-value: 1.20
Transcription factors associated with nkx2.3+nkx2.7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.7
|
ENSDARG00000021232 | NK2 transcription factor related 7 |
|
nkx2.3
|
ENSDARG00000039095 | NK2 homeobox 3 |
|
nkx2.3
|
ENSDARG00000113490 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.7 | dr11_v1_chr8_-_50287949_50287949 | -0.20 | 5.6e-02 | Click! |
| nkx2.3 | dr11_v1_chr13_-_40411908_40411917 | -0.18 | 7.9e-02 | Click! |
Activity profile of nkx2.3+nkx2.7 motif
Sorted Z-values of nkx2.3+nkx2.7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_3828560 | 17.83 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr22_-_13851297 | 14.68 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr20_+_18551657 | 14.31 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr13_+_38430466 | 13.51 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr12_-_25916530 | 13.25 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr15_-_15357178 | 12.81 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr20_-_39271844 | 12.79 |
ENSDART00000192708
|
clu
|
clusterin |
| chr17_+_23298928 | 12.27 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr4_+_21129752 | 11.64 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr14_+_29609245 | 11.41 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr11_+_7324704 | 11.21 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr24_-_6158933 | 11.21 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr24_+_2519761 | 10.70 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr12_+_24342303 | 10.24 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr5_-_58939460 | 9.80 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr1_+_45085194 | 9.78 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr8_+_24861264 | 9.31 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr7_-_28148310 | 9.18 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr17_-_12389259 | 9.13 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr5_+_23622177 | 9.12 |
ENSDART00000121504
|
cx27.5
|
connexin 27.5 |
| chr8_+_31821396 | 9.00 |
ENSDART00000077053
|
plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr21_-_43015383 | 8.90 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr11_+_30057762 | 8.85 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr12_+_34953038 | 8.79 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr1_+_40023640 | 8.75 |
ENSDART00000101623
|
lgi2b
|
leucine-rich repeat LGI family, member 2b |
| chr20_-_28800999 | 8.67 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr18_+_24921587 | 8.21 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr3_+_29714775 | 8.05 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr24_+_34606966 | 7.70 |
ENSDART00000105477
|
lrrtm2
|
leucine rich repeat transmembrane neuronal 2 |
| chr13_+_24834199 | 7.60 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr9_+_40939336 | 7.57 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr4_+_12615836 | 7.55 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr23_-_14990865 | 7.42 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr16_+_32059785 | 7.42 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr3_-_28250722 | 7.40 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr12_+_3022882 | 7.36 |
ENSDART00000122905
|
rac3b
|
Rac family small GTPase 3b |
| chr17_-_6399920 | 7.35 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr12_+_5081759 | 7.28 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr5_-_10946232 | 7.14 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr16_+_20161805 | 7.14 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr17_-_44247707 | 7.01 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr15_-_12319065 | 6.94 |
ENSDART00000162973
ENSDART00000170543 |
fxyd6
|
FXYD domain containing ion transport regulator 6 |
| chr2_+_22702488 | 6.94 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr21_+_28958471 | 6.93 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_-_51323545 | 6.89 |
ENSDART00000139316
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr11_-_6188413 | 6.72 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr7_-_24699985 | 6.71 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr15_-_6247775 | 6.51 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr25_-_7683316 | 6.49 |
ENSDART00000128820
|
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr18_+_40381102 | 6.49 |
ENSDART00000136588
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr10_-_8033468 | 6.45 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr22_+_27090136 | 6.45 |
ENSDART00000136770
|
si:dkey-246e1.3
|
si:dkey-246e1.3 |
| chr22_-_33679277 | 6.41 |
ENSDART00000169948
|
FO904977.1
|
|
| chr16_+_9779680 | 6.38 |
ENSDART00000154253
|
zgc:92912
|
zgc:92912 |
| chr21_+_18353703 | 6.35 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr14_+_25465346 | 6.32 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr24_+_22759451 | 6.18 |
ENSDART00000135392
|
si:dkey-7n6.2
|
si:dkey-7n6.2 |
| chr20_+_34717403 | 6.15 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr5_-_14373662 | 6.12 |
ENSDART00000183694
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr18_+_9637744 | 6.12 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr19_+_24872159 | 6.11 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr23_+_28374458 | 6.10 |
ENSDART00000140058
ENSDART00000144240 |
zgc:153867
|
zgc:153867 |
| chr6_-_41229787 | 6.03 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr23_+_44741500 | 5.98 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr21_+_9628854 | 5.96 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr9_+_21722733 | 5.92 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr9_-_1702648 | 5.86 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr25_-_9805269 | 5.82 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr22_+_18389271 | 5.78 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr15_+_26399538 | 5.72 |
ENSDART00000159589
|
rtn4rl1b
|
reticulon 4 receptor-like 1b |
| chr7_+_32722227 | 5.70 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr9_-_32753535 | 5.69 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr7_-_26497947 | 5.64 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr16_-_55028740 | 5.62 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr2_+_20332044 | 5.49 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr3_-_35800221 | 5.47 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr15_+_36115955 | 5.44 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr2_+_26656283 | 5.44 |
ENSDART00000133202
ENSDART00000099208 |
asph
|
aspartate beta-hydroxylase |
| chr9_-_3671911 | 5.42 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr12_+_7497882 | 5.42 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr6_-_11780070 | 5.41 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr5_-_28679135 | 5.39 |
ENSDART00000193585
|
tnc
|
tenascin C |
| chr19_-_42391383 | 5.35 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr24_-_8729531 | 5.32 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr5_+_32009080 | 5.25 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chr1_-_11973341 | 5.21 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr19_+_42886413 | 5.19 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr8_-_33114202 | 5.14 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr1_-_23110740 | 5.11 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr9_-_18735256 | 5.11 |
ENSDART00000143165
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr5_-_31901468 | 5.10 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr5_+_36611128 | 5.09 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr10_+_37137464 | 5.03 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr8_+_3820134 | 5.03 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr19_+_37848830 | 5.01 |
ENSDART00000042276
ENSDART00000180872 |
nxph1
|
neurexophilin 1 |
| chr23_+_19590006 | 4.99 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr17_+_19630272 | 4.97 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr19_-_44089509 | 4.97 |
ENSDART00000189136
|
rad21b
|
RAD21 cohesin complex component b |
| chr13_-_14487524 | 4.88 |
ENSDART00000141103
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr14_-_24410673 | 4.87 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr1_+_33969015 | 4.86 |
ENSDART00000042984
ENSDART00000146530 |
epha6
|
eph receptor A6 |
| chr3_+_9379877 | 4.85 |
ENSDART00000182080
|
LO018550.1
|
|
| chr21_-_42100471 | 4.84 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr19_-_7358184 | 4.83 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr24_+_41915878 | 4.81 |
ENSDART00000171523
|
TMEM200C
|
transmembrane protein 200C |
| chr10_+_22381802 | 4.78 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr6_-_30210378 | 4.78 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr21_+_34088110 | 4.77 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr23_-_7799184 | 4.76 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr1_-_22512063 | 4.66 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr1_-_50859053 | 4.65 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr14_-_3381303 | 4.62 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr15_-_34213898 | 4.61 |
ENSDART00000191945
ENSDART00000186089 |
etv1
|
ets variant 1 |
| chr15_-_20024205 | 4.55 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr14_-_2206476 | 4.55 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr7_-_28147838 | 4.54 |
ENSDART00000158921
|
lmo1
|
LIM domain only 1 |
| chr19_+_1184878 | 4.53 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr13_-_43599898 | 4.52 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr12_+_39685485 | 4.50 |
ENSDART00000163403
|
LO017650.1
|
|
| chr17_-_42213285 | 4.49 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr12_+_18578597 | 4.48 |
ENSDART00000134944
|
grid2ipb
|
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein, b |
| chr24_-_8729716 | 4.45 |
ENSDART00000183624
|
tfap2a
|
transcription factor AP-2 alpha |
| chr9_+_17983463 | 4.45 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr15_-_31514818 | 4.45 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr3_+_60721342 | 4.42 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr14_-_4682114 | 4.41 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr18_+_10884996 | 4.40 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr10_+_29698467 | 4.38 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr8_+_22582146 | 4.33 |
ENSDART00000157655
ENSDART00000189892 |
CT583651.2
|
|
| chr10_-_8032885 | 4.32 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr9_+_51265283 | 4.30 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr13_+_19322686 | 4.29 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr11_-_27501027 | 4.26 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr14_+_49135264 | 4.24 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr12_+_21298317 | 4.23 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr6_+_40661703 | 4.19 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr5_+_20147830 | 4.16 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr9_+_31534601 | 4.15 |
ENSDART00000133094
|
si:ch211-168k14.2
|
si:ch211-168k14.2 |
| chr7_+_24023653 | 4.11 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr11_+_31323746 | 4.10 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr19_+_26718074 | 4.06 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr22_+_5176255 | 4.05 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr12_+_33038757 | 4.01 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr4_+_12111154 | 3.93 |
ENSDART00000036779
|
tmem178b
|
transmembrane protein 178B |
| chr9_-_33107237 | 3.93 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr8_+_26874924 | 3.93 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr24_+_25069609 | 3.82 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr21_+_22404662 | 3.80 |
ENSDART00000183455
|
lmbrd2b
|
LMBR1 domain containing 2b |
| chr16_-_29480335 | 3.78 |
ENSDART00000148930
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr16_+_39159752 | 3.75 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr12_+_15008582 | 3.73 |
ENSDART00000003847
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr13_+_24662238 | 3.73 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr5_-_12407194 | 3.72 |
ENSDART00000125291
|
ksr2
|
kinase suppressor of ras 2 |
| chr18_-_41375120 | 3.71 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr12_-_8958353 | 3.66 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr19_+_20178978 | 3.63 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr8_-_9570511 | 3.58 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr25_+_14165447 | 3.55 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr19_-_28789404 | 3.54 |
ENSDART00000191453
ENSDART00000026992 |
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr6_+_52350443 | 3.50 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr3_-_30061985 | 3.47 |
ENSDART00000189583
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr18_+_11970987 | 3.45 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr14_-_33894915 | 3.43 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr1_+_7517454 | 3.39 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr19_+_9459050 | 3.39 |
ENSDART00000186419
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr14_+_36628131 | 3.36 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr6_+_37623693 | 3.34 |
ENSDART00000144812
ENSDART00000182709 |
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr6_+_21001264 | 3.33 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr8_-_19467011 | 3.31 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr23_+_37458602 | 3.27 |
ENSDART00000181686
|
cdaa
|
cytidine deaminase a |
| chr24_+_26039464 | 3.27 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr13_+_38990939 | 3.26 |
ENSDART00000145979
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr2_+_39108339 | 3.25 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr8_-_50888806 | 3.21 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr11_+_30244356 | 3.20 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr5_+_32206378 | 3.20 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr20_-_23171430 | 3.13 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr14_+_3449780 | 3.11 |
ENSDART00000163849
|
trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr16_-_13680692 | 3.11 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr16_+_46459680 | 3.11 |
ENSDART00000101698
|
rpz3
|
rapunzel 3 |
| chr22_-_3564563 | 3.10 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr19_+_13099541 | 3.10 |
ENSDART00000171607
ENSDART00000165448 ENSDART00000170365 |
rims2b
|
regulating synaptic membrane exocytosis 2b |
| chr8_-_19051906 | 3.09 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr14_-_2203124 | 3.08 |
ENSDART00000124171
|
si:dkeyp-115d7.2
|
si:dkeyp-115d7.2 |
| chr20_-_35052823 | 3.08 |
ENSDART00000153033
|
kif26bb
|
kinesin family member 26Bb |
| chr24_+_11334733 | 3.08 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr2_+_30786773 | 3.08 |
ENSDART00000019029
ENSDART00000145681 |
atp6v1h
|
ATPase H+ transporting V1 subunit H |
| chr1_-_13271569 | 3.05 |
ENSDART00000127838
|
pcdh18a
|
protocadherin 18a |
| chr1_-_13271085 | 3.02 |
ENSDART00000193663
|
pcdh18a
|
protocadherin 18a |
| chr11_+_44804685 | 3.02 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr17_-_14700889 | 3.01 |
ENSDART00000179975
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr21_+_28478663 | 2.95 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr10_+_33744098 | 2.94 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr7_+_73397283 | 2.85 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr10_+_42690374 | 2.83 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr5_-_67145505 | 2.83 |
ENSDART00000011295
|
rom1a
|
retinal outer segment membrane protein 1a |
| chr2_-_32352946 | 2.82 |
ENSDART00000144870
ENSDART00000077151 |
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr20_-_47732703 | 2.80 |
ENSDART00000193975
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr17_-_3986236 | 2.78 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr5_+_32009542 | 2.75 |
ENSDART00000182025
ENSDART00000179879 |
scai
|
suppressor of cancer cell invasion |
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.3+nkx2.7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 29.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 3.1 | 9.3 | GO:0015824 | proline transport(GO:0015824) |
| 2.7 | 13.5 | GO:0016322 | neuron remodeling(GO:0016322) |
| 2.2 | 11.2 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 2.1 | 6.2 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.9 | 7.6 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 1.8 | 5.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 1.7 | 6.7 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 1.6 | 8.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 1.4 | 5.8 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 1.4 | 5.7 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 1.3 | 5.2 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 1.3 | 6.3 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 1.2 | 4.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.1 | 5.5 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 1.1 | 3.3 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 1.1 | 5.4 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.1 | 4.2 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 1.0 | 4.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.9 | 3.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.9 | 5.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.9 | 2.7 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.9 | 4.4 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.8 | 5.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.8 | 10.8 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.8 | 16.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.7 | 8.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.7 | 3.6 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.7 | 6.1 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.7 | 2.0 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) regulation of phospholipase C activity(GO:1900274) |
| 0.6 | 9.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.6 | 8.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.6 | 10.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.6 | 2.3 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.6 | 13.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.5 | 4.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.5 | 7.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.5 | 3.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.5 | 5.0 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.5 | 5.0 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.5 | 1.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.5 | 7.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.5 | 6.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.5 | 6.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.4 | 7.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.4 | 1.3 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.4 | 7.4 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.4 | 2.0 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.4 | 2.7 | GO:0070206 | protein trimerization(GO:0070206) protein homotrimerization(GO:0070207) |
| 0.4 | 2.2 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.3 | 1.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.3 | 2.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 6.2 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.3 | 1.9 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.3 | 6.0 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.3 | 1.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.3 | 6.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 3.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 6.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 2.0 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 17.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.3 | 2.2 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 1.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.3 | 1.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.3 | 8.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 1.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.3 | 1.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 1.0 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.3 | 4.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 1.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 2.9 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.2 | 4.4 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.2 | 0.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.2 | 3.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 29.3 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.2 | 4.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.2 | 9.8 | GO:0071696 | ectodermal placode development(GO:0071696) |
| 0.2 | 0.7 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.2 | 3.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 3.1 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 10.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.2 | 10.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.2 | 2.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.1 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.2 | 6.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 4.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 1.0 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.2 | 0.6 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 10.7 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.2 | 6.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 1.5 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.2 | 1.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.5 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.2 | 0.9 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 10.2 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.2 | 9.0 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.2 | 3.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.7 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 1.0 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 6.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.7 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.7 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 3.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 3.8 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 3.8 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.1 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.7 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 9.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.8 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 1.7 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.1 | 2.3 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 9.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 2.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 11.4 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.1 | 6.6 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 2.7 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.1 | 1.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.8 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 1.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 3.2 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 1.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.8 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 3.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 14.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 2.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 2.0 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 2.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.7 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 2.1 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 2.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.6 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 1.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 7.7 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 2.1 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 1.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.8 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 3.6 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 2.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.9 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.2 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 2.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 3.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 3.5 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.2 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 3.7 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 1.6 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.0 | 0.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 1.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 8.1 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 0.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 4.1 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 4.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 4.0 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 2.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 2.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.7 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 2.0 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.7 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 24.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 2.3 | 13.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.6 | 4.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.5 | 9.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.2 | 6.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.1 | 10.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.8 | 3.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.8 | 8.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.8 | 5.5 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.7 | 5.0 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.7 | 26.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.6 | 6.0 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.6 | 2.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.6 | 3.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 4.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 5.0 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.5 | 1.9 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.5 | 7.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 10.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 2.0 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 1.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 1.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 1.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 5.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 14.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.3 | 4.1 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 0.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.3 | 9.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 1.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.3 | 5.0 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 10.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 2.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 3.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 2.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 2.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 14.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 8.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.2 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 3.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 1.0 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 3.1 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.0 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 11.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 2.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 6.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 7.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 6.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 2.1 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.0 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 13.5 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 8.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 1.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 16.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 2.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 4.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.7 | 29.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 4.4 | 13.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 3.7 | 14.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 2.1 | 6.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.4 | 5.4 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 1.3 | 3.9 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 1.2 | 6.9 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.0 | 5.9 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.9 | 5.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.9 | 4.3 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.8 | 13.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.8 | 3.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.8 | 5.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.7 | 8.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.7 | 10.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.7 | 3.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.7 | 2.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.7 | 9.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.6 | 25.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.6 | 6.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 2.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.6 | 4.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.6 | 5.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.5 | 4.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 2.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.5 | 3.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.5 | 6.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.5 | 1.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.5 | 4.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 6.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.5 | 3.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.5 | 12.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 6.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 8.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.4 | 7.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.4 | 10.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 2.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 2.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 5.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.3 | 1.9 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 4.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 11.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 6.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 3.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 9.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 2.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.3 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 5.0 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.3 | 5.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.2 | 1.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 2.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 11.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 1.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 4.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 3.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 18.3 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.2 | 3.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 3.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 5.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 1.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 3.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 5.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 0.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 1.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 11.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 6.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 11.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 4.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 7.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.2 | 3.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 3.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 11.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 4.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 4.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 7.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 2.8 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.8 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 2.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 4.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.6 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 3.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.0 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 3.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.7 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 4.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 10.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 9.0 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 21.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 14.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 7.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 11.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 3.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 3.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 2.9 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 3.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 10.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.3 | 5.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 4.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 6.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 13.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 11.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 3.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 2.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 2.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 4.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 6.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 3.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 7.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 2.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 4.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 12.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 14.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 1.1 | 12.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.8 | 11.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.8 | 3.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.7 | 6.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.7 | 7.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.6 | 9.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 4.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.4 | 6.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 10.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 2.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 2.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 3.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 2.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 4.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 1.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 3.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 1.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 3.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 2.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 4.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.2 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 4.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 5.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |