Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
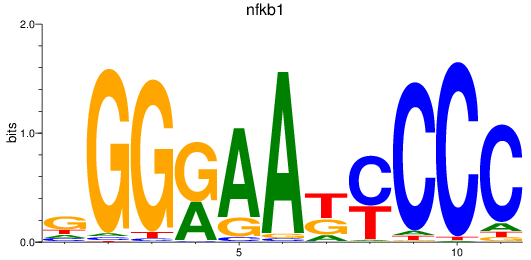
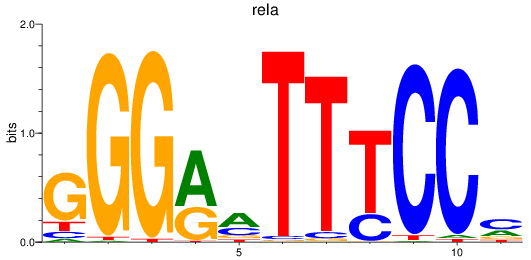
Results for nfkb1_rela
Z-value: 1.62
Transcription factors associated with nfkb1_rela
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfkb1
|
ENSDARG00000105261 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
|
rela
|
ENSDARG00000098696 | v-rel avian reticuloendotheliosis viral oncogene homolog A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfkb1 | dr11_v1_chr14_-_49859747_49859823 | 0.49 | 6.0e-07 | Click! |
| rela | dr11_v1_chr7_-_804515_804515 | 0.30 | 3.0e-03 | Click! |
Activity profile of nfkb1_rela motif
Sorted Z-values of nfkb1_rela motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_18716 | 21.53 |
ENSDART00000172454
ENSDART00000161190 |
nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr24_+_26006730 | 21.12 |
ENSDART00000140384
ENSDART00000139184 |
ccl20b
|
chemokine (C-C motif) ligand 20b |
| chr20_+_16881883 | 19.49 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr20_-_51307815 | 19.06 |
ENSDART00000098833
|
nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr2_+_42191592 | 17.31 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr7_+_12950507 | 16.77 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr8_+_24281512 | 16.09 |
ENSDART00000062845
|
mmp9
|
matrix metallopeptidase 9 |
| chr1_-_37377509 | 16.01 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr1_-_52437056 | 13.78 |
ENSDART00000138337
|
si:ch211-217k17.12
|
si:ch211-217k17.12 |
| chr7_-_34262080 | 13.65 |
ENSDART00000183246
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr5_+_22579975 | 13.44 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr1_-_52431220 | 13.29 |
ENSDART00000111256
|
zgc:194101
|
zgc:194101 |
| chr12_+_20641471 | 13.10 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr18_-_5527050 | 13.03 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr8_-_18667693 | 12.84 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr13_-_25819825 | 12.31 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr23_+_31815423 | 12.27 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_-_21242460 | 12.21 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr15_+_32727848 | 12.14 |
ENSDART00000161361
|
postnb
|
periostin, osteoblast specific factor b |
| chr12_+_22576404 | 12.00 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr24_-_33703504 | 11.40 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr12_+_20641102 | 11.23 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr4_-_12795436 | 11.17 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr17_+_12942634 | 10.33 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr21_+_19547806 | 10.26 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr9_-_51563575 | 9.95 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr8_-_12432604 | 9.73 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr23_+_19701587 | 9.72 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr16_+_23403602 | 9.52 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr16_-_29714540 | 9.36 |
ENSDART00000067854
|
tnfaip8l2b
|
tumor necrosis factor, alpha-induced protein 8-like 2b |
| chr14_+_3038473 | 9.33 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr19_+_7124337 | 9.33 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr1_-_49225890 | 9.33 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr6_+_54576520 | 9.28 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr14_-_4121052 | 9.22 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr6_+_3334710 | 9.15 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr19_-_7144548 | 9.09 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr24_+_22731228 | 8.78 |
ENSDART00000146733
|
si:dkey-225k4.1
|
si:dkey-225k4.1 |
| chr6_-_436658 | 8.75 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr14_-_4120636 | 8.68 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr9_+_41156287 | 8.68 |
ENSDART00000189195
ENSDART00000186270 |
stat4
|
signal transducer and activator of transcription 4 |
| chr23_-_32162810 | 8.59 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr5_-_42878178 | 8.54 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr9_-_98982 | 8.32 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr24_+_25919809 | 8.07 |
ENSDART00000006615
|
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr14_-_1646360 | 8.01 |
ENSDART00000186528
|
LO018106.2
|
|
| chr2_+_45191049 | 7.95 |
ENSDART00000165392
|
ccl20a.3
|
chemokine (C-C motif) ligand 20a, duplicate 3 |
| chr20_+_23501535 | 7.94 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr21_-_22693730 | 7.93 |
ENSDART00000158342
|
gig2d
|
grass carp reovirus (GCRV)-induced gene 2d |
| chr6_+_2030703 | 7.82 |
ENSDART00000109679
ENSDART00000187502 ENSDART00000191165 ENSDART00000187544 |
quo
|
quattro |
| chr18_-_11729 | 7.77 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr9_+_41156818 | 7.63 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr22_-_651719 | 7.39 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr12_-_30760971 | 7.31 |
ENSDART00000066257
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr21_+_30937690 | 7.13 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr21_-_435466 | 6.87 |
ENSDART00000110297
|
klf4
|
Kruppel-like factor 4 |
| chr5_+_13373593 | 6.80 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr11_+_8129536 | 6.66 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr23_-_12423778 | 6.56 |
ENSDART00000124091
|
wfdc2
|
WAP four-disulfide core domain 2 |
| chr23_-_38054 | 6.50 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr23_+_42819221 | 6.36 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr13_+_32148338 | 6.33 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr2_-_51772438 | 6.27 |
ENSDART00000170241
|
BX908782.2
|
Danio rerio three-finger protein 5 (LOC100003647), mRNA. |
| chr8_-_13029297 | 6.24 |
ENSDART00000144305
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr24_+_80653 | 6.17 |
ENSDART00000158473
ENSDART00000129135 |
reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr6_+_3334392 | 6.17 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr1_-_52447364 | 6.03 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr8_+_25302172 | 5.99 |
ENSDART00000046182
ENSDART00000145316 |
gstm.3
|
glutathione S-transferase mu tandem duplicate 3 |
| chr19_+_19767567 | 5.89 |
ENSDART00000169074
|
hoxa3a
|
homeobox A3a |
| chr19_+_7043634 | 5.88 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr16_-_25739331 | 5.84 |
ENSDART00000189455
|
bcl3
|
B cell CLL/lymphoma 3 |
| chr5_+_24086227 | 5.83 |
ENSDART00000051549
ENSDART00000177458 ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr4_-_149334 | 5.82 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr1_-_52443379 | 5.78 |
ENSDART00000144676
|
si:ch211-217k17.11
|
si:ch211-217k17.11 |
| chr7_+_29293452 | 5.76 |
ENSDART00000127358
|
si:ch211-112g6.4
|
si:ch211-112g6.4 |
| chr13_+_22717366 | 5.75 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr6_-_442163 | 5.70 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr16_+_38394371 | 5.55 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr16_-_35975254 | 5.55 |
ENSDART00000167537
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr5_+_9246018 | 5.44 |
ENSDART00000081769
ENSDART00000183189 |
susd1
|
sushi domain containing 1 |
| chr23_+_10347851 | 5.25 |
ENSDART00000127667
|
krt18
|
keratin 18 |
| chr11_+_6456146 | 5.23 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr25_-_13188214 | 5.17 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr3_-_32170850 | 5.11 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr4_+_70151160 | 5.04 |
ENSDART00000111816
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr17_-_29249258 | 5.04 |
ENSDART00000031458
|
traf3
|
TNF receptor-associated factor 3 |
| chr17_+_51746830 | 4.96 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr7_-_22632518 | 4.94 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr5_+_13385837 | 4.93 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr9_-_6380653 | 4.92 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr7_-_22632690 | 4.87 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr14_+_36885524 | 4.85 |
ENSDART00000032547
|
lect2l
|
leukocyte cell-derived chemotaxin 2 like |
| chr2_-_2642476 | 4.80 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr9_-_23894392 | 4.77 |
ENSDART00000133417
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr14_+_36889893 | 4.77 |
ENSDART00000124159
|
si:ch211-132p1.3
|
si:ch211-132p1.3 |
| chr13_-_51846224 | 4.74 |
ENSDART00000184663
|
LT631684.2
|
|
| chr13_+_30692669 | 4.74 |
ENSDART00000187818
|
CR762483.1
|
|
| chr6_+_18298444 | 4.67 |
ENSDART00000166018
|
card14
|
caspase recruitment domain family, member 14 |
| chr25_-_6557854 | 4.64 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr13_+_22717939 | 4.61 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr17_-_7733037 | 4.59 |
ENSDART00000064657
|
stx11a
|
syntaxin 11a |
| chr7_+_57677120 | 4.55 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr11_-_25853212 | 4.43 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr1_+_27690 | 4.38 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr1_+_33328857 | 4.27 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr3_+_57820913 | 4.26 |
ENSDART00000168101
|
CU571328.1
|
|
| chr9_-_32604414 | 4.19 |
ENSDART00000088876
ENSDART00000166502 |
satb2
|
SATB homeobox 2 |
| chr1_-_59232267 | 4.18 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr13_-_39947335 | 4.18 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr19_+_46113828 | 4.16 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr14_-_49859747 | 4.16 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr15_-_4152582 | 4.16 |
ENSDART00000171942
|
si:dkey-83h2.5
|
si:dkey-83h2.5 |
| chr2_+_36004381 | 4.12 |
ENSDART00000098706
|
lamc2
|
laminin, gamma 2 |
| chr15_+_27364394 | 4.09 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr2_+_21356242 | 4.03 |
ENSDART00000145670
ENSDART00000146600 |
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr22_+_16497670 | 4.02 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr12_+_10631266 | 4.01 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr7_+_19835569 | 3.98 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr7_-_59123066 | 3.92 |
ENSDART00000175438
|
dennd4c
|
DENN/MADD domain containing 4C |
| chr7_-_20241346 | 3.92 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr23_-_33738570 | 3.90 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr14_-_30967284 | 3.83 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr6_-_54815886 | 3.78 |
ENSDART00000180793
ENSDART00000007498 |
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr6_-_52675630 | 3.78 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr11_+_14333441 | 3.78 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr22_+_661711 | 3.65 |
ENSDART00000113795
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr14_+_30773764 | 3.63 |
ENSDART00000186961
|
atl3
|
atlastin 3 |
| chr12_+_48634927 | 3.62 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr19_-_10432134 | 3.58 |
ENSDART00000081440
|
il11b
|
interleukin 11b |
| chr8_+_22355909 | 3.54 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr6_+_49771626 | 3.53 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr14_+_49376011 | 3.47 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr7_-_8309505 | 3.44 |
ENSDART00000182530
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr6_+_19933763 | 3.40 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr22_-_31517300 | 3.38 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr19_+_43119698 | 3.36 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr8_-_20230559 | 3.33 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr3_+_22273123 | 3.32 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr12_+_46740584 | 3.28 |
ENSDART00000171563
|
plaub
|
plasminogen activator, urokinase b |
| chr14_-_6987649 | 3.25 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr2_-_45191319 | 3.23 |
ENSDART00000192272
|
CR407590.2
|
|
| chr23_-_437467 | 3.14 |
ENSDART00000192106
|
tspan2b
|
tetraspanin 2b |
| chr2_-_53592532 | 3.12 |
ENSDART00000184066
|
ccl25a
|
chemokine (C-C motif) ligand 25a |
| chr1_+_33322555 | 3.12 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
| chr25_+_8955530 | 3.12 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr7_-_73843720 | 3.11 |
ENSDART00000111622
|
caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr17_+_8175998 | 3.09 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr16_+_35535375 | 3.05 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr6_+_49771372 | 3.04 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr10_+_8875195 | 3.04 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr22_+_8536838 | 3.02 |
ENSDART00000132998
|
si:ch73-27e22.7
|
si:ch73-27e22.7 |
| chr17_-_2685026 | 2.95 |
ENSDART00000191014
ENSDART00000179309 |
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr21_+_15866522 | 2.95 |
ENSDART00000110728
|
gcnt4b
|
glucosaminyl (N-acetyl) transferase 4, core 2, b |
| chr4_-_12914163 | 2.94 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr3_+_59899452 | 2.92 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr7_-_20338048 | 2.87 |
ENSDART00000125594
|
zgc:194312
|
zgc:194312 |
| chr12_+_27462225 | 2.85 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr7_-_22632938 | 2.84 |
ENSDART00000159867
ENSDART00000165706 |
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr18_-_18543358 | 2.83 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr7_-_51461649 | 2.82 |
ENSDART00000193947
ENSDART00000174328 |
arhgap36
|
Rho GTPase activating protein 36 |
| chr14_+_16765992 | 2.77 |
ENSDART00000140061
|
sqstm1
|
sequestosome 1 |
| chr16_+_34111919 | 2.76 |
ENSDART00000134037
ENSDART00000006061 ENSDART00000140552 |
tcea3
|
transcription elongation factor A (SII), 3 |
| chr22_-_10354381 | 2.75 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr10_+_20608676 | 2.70 |
ENSDART00000140141
|
si:dkey-81j8.6
|
si:dkey-81j8.6 |
| chr7_-_804515 | 2.68 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr5_-_44496553 | 2.67 |
ENSDART00000178081
|
gas1a
|
growth arrest-specific 1a |
| chr16_-_27749172 | 2.64 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr5_+_9246458 | 2.62 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr21_+_45626136 | 2.60 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr8_+_25629722 | 2.59 |
ENSDART00000026807
|
wasa
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) a |
| chr7_-_28696556 | 2.57 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr12_-_3903886 | 2.56 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr4_-_75681004 | 2.53 |
ENSDART00000186296
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr7_-_22941472 | 2.53 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr10_+_31646020 | 2.53 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr4_+_76789395 | 2.51 |
ENSDART00000147574
|
si:ch73-56d11.3
|
si:ch73-56d11.3 |
| chr25_+_16646113 | 2.50 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr12_+_46745239 | 2.50 |
ENSDART00000057179
|
plaub
|
plasminogen activator, urokinase b |
| chr4_-_22311610 | 2.48 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr12_-_11349899 | 2.45 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
| chr19_+_20713424 | 2.45 |
ENSDART00000129730
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr22_+_19425657 | 2.45 |
ENSDART00000187531
ENSDART00000078804 |
si:dkey-78l4.5
|
si:dkey-78l4.5 |
| chr5_-_44496805 | 2.44 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr4_+_68562464 | 2.43 |
ENSDART00000192954
|
BX548011.4
|
|
| chr7_+_34786591 | 2.42 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr7_+_25053331 | 2.42 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr20_-_39367895 | 2.41 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr20_+_218886 | 2.41 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr15_+_17251191 | 2.40 |
ENSDART00000156587
|
si:ch73-223p23.2
|
si:ch73-223p23.2 |
| chr5_+_51909740 | 2.37 |
ENSDART00000162541
|
thbs4a
|
thrombospondin 4a |
| chr1_+_17892944 | 2.37 |
ENSDART00000013021
|
tlr3
|
toll-like receptor 3 |
| chr2_-_879800 | 2.34 |
ENSDART00000019733
|
irf4a
|
interferon regulatory factor 4a |
| chr11_+_35050253 | 2.32 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr14_-_40850481 | 2.32 |
ENSDART00000173236
|
elf1
|
E74-like ETS transcription factor 1 |
| chr4_+_77681389 | 2.29 |
ENSDART00000099727
|
gbp4
|
guanylate binding protein 4 |
| chr5_+_37785152 | 2.27 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr2_-_57227542 | 2.26 |
ENSDART00000182675
ENSDART00000159480 |
btbd2b
|
BTB (POZ) domain containing 2b |
| chr5_-_64168415 | 2.20 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr24_+_41273252 | 2.19 |
ENSDART00000105526
|
nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr23_-_29003864 | 2.19 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfkb1_rela
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.5 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 6.1 | 18.4 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 5.4 | 16.1 | GO:0002631 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 5.0 | 14.9 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 3.3 | 20.0 | GO:0051883 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 3.1 | 12.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 2.5 | 7.4 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 2.1 | 6.3 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 2.1 | 6.2 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 2.0 | 12.3 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 1.9 | 5.8 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 1.6 | 4.9 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.6 | 11.4 | GO:0033292 | T-tubule organization(GO:0033292) |
| 1.4 | 2.8 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 1.4 | 4.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.4 | 4.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 1.4 | 4.1 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 1.2 | 7.3 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 1.0 | 8.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 1.0 | 3.1 | GO:0072679 | thymocyte migration(GO:0072679) |
| 1.0 | 40.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 1.0 | 8.6 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.9 | 3.8 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.9 | 2.8 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.9 | 14.5 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.8 | 5.9 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.8 | 2.5 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.8 | 3.2 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.8 | 11.2 | GO:0034122 | regulation of toll-like receptor signaling pathway(GO:0034121) negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.8 | 14.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.7 | 2.9 | GO:0030091 | protein repair(GO:0030091) |
| 0.7 | 2.9 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.7 | 5.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 15.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.7 | 2.8 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.7 | 10.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.7 | 2.6 | GO:0015677 | copper ion import(GO:0015677) |
| 0.6 | 13.9 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.6 | 2.3 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.6 | 5.8 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.6 | 9.7 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.6 | 4.0 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.6 | 5.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.5 | 2.2 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.5 | 1.6 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.5 | 8.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.5 | 1.6 | GO:0007567 | parturition(GO:0007567) maternal process involved in parturition(GO:0060137) |
| 0.5 | 2.6 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.5 | 1.5 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.5 | 1.9 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 1.3 | GO:0060879 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.4 | 4.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.4 | 2.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.4 | 2.0 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.4 | 1.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.4 | 1.6 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.4 | 12.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.4 | 2.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.3 | 10.7 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.3 | 1.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 14.7 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.3 | 9.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.3 | 3.5 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.3 | 2.8 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.3 | 9.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.3 | 1.7 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.3 | 7.7 | GO:0072089 | stem cell proliferation(GO:0072089) |
| 0.3 | 3.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 2.5 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 5.1 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.2 | 1.9 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.2 | 2.6 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.2 | 3.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.2 | 0.9 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 6.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.2 | 3.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.2 | 3.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.6 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.2 | 0.8 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.2 | 18.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.2 | 2.6 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 6.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.2 | 0.6 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.2 | 13.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.2 | 1.7 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 10.7 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 6.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 2.9 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 5.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 1.1 | GO:0003303 | BMP signaling pathway involved in determination of left/right symmetry(GO:0003154) BMP signaling pathway involved in heart jogging(GO:0003303) locus ceruleus development(GO:0021703) |
| 0.1 | 2.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 7.4 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.1 | 1.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.5 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 3.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.8 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 4.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 1.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 1.7 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 0.4 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 3.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 15.9 | GO:0045087 | innate immune response(GO:0045087) |
| 0.1 | 4.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 2.8 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.1 | 0.3 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.1 | 4.2 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.1 | 0.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 1.0 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 5.8 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.1 | 1.0 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 1.8 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 3.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 2.8 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.1 | 2.9 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 1.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 4.3 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 1.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 2.0 | GO:0001816 | cytokine production(GO:0001816) |
| 0.0 | 1.9 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.9 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 2.0 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 5.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 7.6 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.0 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 1.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.4 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 6.1 | GO:0051603 | cellular protein catabolic process(GO:0044257) proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 2.0 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 1.0 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 6.0 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.7 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.4 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 1.5 | GO:0040008 | regulation of growth(GO:0040008) |
| 0.0 | 4.8 | GO:0008283 | cell proliferation(GO:0008283) |
| 0.0 | 1.7 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 7.3 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.3 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.5 | 6.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 1.2 | 9.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.1 | 16.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.9 | 13.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.7 | 28.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.6 | 2.5 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.6 | 2.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.5 | 3.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 9.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.3 | 4.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 4.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 6.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 3.4 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.2 | 2.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 8.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 1.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 8.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 12.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 13.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.4 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 18.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 5.6 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.1 | 13.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 5.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 12.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 6.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 15.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 7.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 46.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 3.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 2.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 4.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 5.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 73.5 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 13.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 2.2 | 15.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 2.1 | 12.6 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.4 | 21.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 1.4 | 4.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.3 | 6.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 1.1 | 7.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.0 | 43.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 1.0 | 9.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.9 | 31.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.8 | 8.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.7 | 2.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.7 | 2.0 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.7 | 2.6 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.6 | 2.9 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.6 | 2.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.6 | 5.0 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.5 | 1.6 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.5 | 12.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.5 | 2.9 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.5 | 9.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.5 | 6.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 4.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.5 | 13.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.5 | 3.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.4 | 4.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 12.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.4 | 7.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.4 | 9.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 1.6 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.4 | 2.9 | GO:0004984 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.4 | 8.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 9.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 3.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 5.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 3.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 5.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 6.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 1.5 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 0.5 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 3.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 7.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 4.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 4.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 18.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 3.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.4 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 3.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.1 | 0.8 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 4.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 6.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 3.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 7.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 9.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 13.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 1.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.9 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 21.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.6 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.7 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 9.2 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.1 | 11.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 0.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 16.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 6.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 2.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 12.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 52.6 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.0 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.3 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 2.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 18.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 1.1 | 21.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 1.0 | 19.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.8 | 24.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.7 | 13.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.7 | 19.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.6 | 12.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.5 | 4.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 8.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 5.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 2.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 9.9 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 5.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.2 | 8.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 2.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 2.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 2.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 4.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 4.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 6.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 0.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 2.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 6.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 5.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 5.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 2.3 | 16.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 2.2 | 11.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 1.9 | 33.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 1.8 | 10.7 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 1.2 | 5.0 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.8 | 2.4 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.8 | 5.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.6 | 17.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.6 | 5.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.6 | 6.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 24.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.5 | 17.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.4 | 8.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.4 | 4.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 8.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.3 | 0.9 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 6.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 1.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.2 | 3.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 11.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 4.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 2.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.9 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |