Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
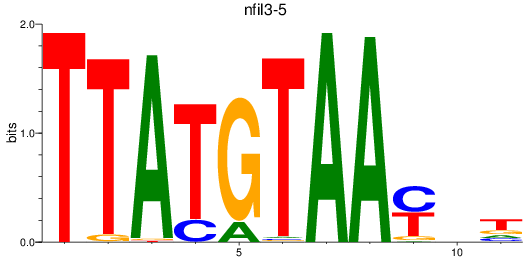
Results for nfil3-5
Z-value: 1.45
Transcription factors associated with nfil3-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfil3-5
|
ENSDARG00000094965 | nuclear factor, interleukin 3 regulated, member 5 |
|
nfil3-5
|
ENSDARG00000113345 | nuclear factor, interleukin 3 regulated, member 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfil3-5 | dr11_v1_chr6_+_8172227_8172227 | 0.57 | 2.8e-09 | Click! |
Activity profile of nfil3-5 motif
Sorted Z-values of nfil3-5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_34984333 | 32.29 |
ENSDART00000154760
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr21_-_22114625 | 25.89 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr5_+_37837245 | 25.46 |
ENSDART00000171617
|
epd
|
ependymin |
| chr24_-_38079261 | 23.47 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr17_-_43466317 | 23.12 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr3_-_46818001 | 22.93 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr14_+_33458294 | 22.09 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr3_+_16612574 | 22.07 |
ENSDART00000104481
|
slc17a7a
|
solute carrier family 17 (vesicular glutamate transporter), member 7a |
| chr17_-_20979077 | 19.73 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr10_-_24371312 | 19.35 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr7_+_30787903 | 18.81 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr21_-_22115136 | 18.80 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr4_-_5302866 | 18.48 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr21_-_42202792 | 17.83 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr7_+_38717624 | 17.57 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr3_-_35602233 | 17.26 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr25_+_15354095 | 16.95 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr23_+_45579497 | 16.24 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr23_+_37323962 | 16.05 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr25_-_11088839 | 15.79 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr13_-_21688176 | 15.73 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr2_+_24177190 | 15.42 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr16_+_46111849 | 15.24 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr1_-_14234076 | 14.69 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr16_+_39146696 | 14.61 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr21_-_43949208 | 14.45 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr13_+_27314795 | 14.42 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr21_-_43550120 | 14.38 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr1_-_14233815 | 14.35 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr19_+_23982466 | 14.19 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr1_-_8101495 | 14.18 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr3_-_28665291 | 13.78 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr11_-_10770053 | 13.06 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr6_-_13187168 | 12.92 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr19_-_5254699 | 12.85 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr6_+_12326267 | 12.75 |
ENSDART00000155101
|
si:dkey-276j7.3
|
si:dkey-276j7.3 |
| chr4_-_5302162 | 12.11 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr7_+_20535869 | 11.83 |
ENSDART00000078181
|
zgc:158423
|
zgc:158423 |
| chr5_-_55848358 | 11.74 |
ENSDART00000130891
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr14_-_21219659 | 11.74 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr5_-_55848511 | 11.65 |
ENSDART00000183503
|
camk4
|
calcium/calmodulin-dependent protein kinase IV |
| chr10_-_26729930 | 11.56 |
ENSDART00000145532
|
fgf13b
|
fibroblast growth factor 13b |
| chr5_-_21970881 | 11.22 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr5_+_1278092 | 11.08 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr14_-_21618005 | 11.04 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr8_-_32497815 | 10.71 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr6_+_41255485 | 10.66 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr18_-_21859019 | 10.60 |
ENSDART00000100885
|
nrn1la
|
neuritin 1-like a |
| chr1_-_53750522 | 10.57 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr4_-_22671469 | 10.47 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr25_-_10564721 | 10.35 |
ENSDART00000154776
|
galn
|
galanin/GMAP prepropeptide |
| chr10_+_21776911 | 10.20 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr9_-_18877597 | 10.11 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr2_+_34967022 | 10.10 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr10_-_26738209 | 10.01 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr4_-_5291256 | 9.80 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr18_-_1228688 | 9.79 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr23_+_17800717 | 9.66 |
ENSDART00000122654
ENSDART00000044986 |
rnd1a
|
Rho family GTPase 1a |
| chr8_-_32497581 | 9.40 |
ENSDART00000176298
ENSDART00000183340 |
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr2_+_34967210 | 9.26 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr6_-_35472923 | 9.14 |
ENSDART00000185907
|
rgs8
|
regulator of G protein signaling 8 |
| chr5_+_29784172 | 8.93 |
ENSDART00000139035
|
si:ch211-215c18.3
|
si:ch211-215c18.3 |
| chr3_+_33345348 | 8.85 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr23_+_19590006 | 8.77 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr25_+_7784582 | 8.68 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr5_-_22001003 | 8.66 |
ENSDART00000134393
ENSDART00000143878 |
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr12_-_26383242 | 8.63 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr3_+_37827373 | 8.54 |
ENSDART00000039517
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr7_-_32782430 | 8.50 |
ENSDART00000173808
|
gas2b
|
growth arrest-specific 2b |
| chr8_+_47897734 | 8.45 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr25_-_10565006 | 8.40 |
ENSDART00000130608
ENSDART00000190212 |
galn
|
galanin/GMAP prepropeptide |
| chr7_-_32833153 | 8.39 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr14_-_9199968 | 8.14 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr3_+_15296824 | 8.07 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr19_+_7549854 | 8.03 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr25_-_9805269 | 8.01 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr5_-_40210447 | 7.92 |
ENSDART00000131323
|
si:dkey-193c22.1
|
si:dkey-193c22.1 |
| chr20_-_40717900 | 7.89 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr18_+_1703984 | 7.86 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr11_+_30161699 | 7.83 |
ENSDART00000190504
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr1_+_31864404 | 7.81 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr7_-_38861741 | 7.73 |
ENSDART00000173629
ENSDART00000037361 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr9_+_17862858 | 7.73 |
ENSDART00000166566
|
dgkh
|
diacylglycerol kinase, eta |
| chr9_-_18742704 | 7.58 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr9_+_46644633 | 7.45 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr7_+_26629084 | 7.41 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr22_-_354592 | 7.37 |
ENSDART00000155769
|
tmem240b
|
transmembrane protein 240b |
| chr18_-_898870 | 7.27 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr10_-_17284055 | 7.26 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr4_+_3482312 | 7.13 |
ENSDART00000109044
|
grm8a
|
glutamate receptor, metabotropic 8a |
| chr20_+_1121458 | 6.96 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr14_-_18671334 | 6.90 |
ENSDART00000182381
|
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr9_+_17429170 | 6.87 |
ENSDART00000006256
|
zgc:101559
|
zgc:101559 |
| chr6_-_12314475 | 6.81 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr13_-_27675212 | 6.81 |
ENSDART00000141035
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr19_-_28367413 | 6.77 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr14_+_36628131 | 6.68 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr24_-_17023392 | 6.67 |
ENSDART00000106058
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr6_+_8172227 | 6.58 |
ENSDART00000146106
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr15_-_9272328 | 6.56 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr19_-_12145765 | 6.49 |
ENSDART00000032474
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr23_-_5032587 | 6.46 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr21_-_42007213 | 6.44 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr6_-_21616659 | 6.28 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr5_-_41494831 | 6.28 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr7_+_38278860 | 6.19 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr4_-_40972176 | 6.05 |
ENSDART00000152052
|
si:dkeyp-82h4.1
|
si:dkeyp-82h4.1 |
| chr11_+_6819050 | 6.03 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr22_+_786556 | 6.01 |
ENSDART00000125347
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr15_-_26636826 | 6.01 |
ENSDART00000087632
|
slc47a4
|
solute carrier family 47 (multidrug and toxin extrusion), member 4 |
| chr1_+_54673846 | 5.97 |
ENSDART00000145018
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr16_-_21047872 | 5.88 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr9_-_18743012 | 5.81 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr12_+_36416173 | 5.74 |
ENSDART00000190278
|
unk
|
unkempt family zinc finger |
| chr21_-_12119711 | 5.69 |
ENSDART00000131538
|
celf4
|
CUGBP, Elav-like family member 4 |
| chr20_-_44576949 | 5.66 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr5_-_30704390 | 5.64 |
ENSDART00000016709
|
ift22
|
intraflagellar transport 22 homolog (Chlamydomonas) |
| chr15_+_7992906 | 5.61 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr6_-_29612269 | 5.54 |
ENSDART00000104293
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr11_+_30161168 | 5.51 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr16_-_45069882 | 5.50 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr5_-_50638905 | 5.49 |
ENSDART00000180842
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr22_-_3595439 | 5.48 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr19_-_12145390 | 5.44 |
ENSDART00000143087
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr20_+_38201644 | 5.40 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr6_-_18121075 | 5.40 |
ENSDART00000171072
|
SEC14L1
|
si:dkey-237i9.1 |
| chr3_-_21348478 | 5.40 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr8_-_39903803 | 5.37 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr18_+_17428506 | 5.33 |
ENSDART00000100223
|
zgc:91860
|
zgc:91860 |
| chr19_-_41371978 | 5.33 |
ENSDART00000166063
ENSDART00000170343 |
slc25a13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr5_+_38276582 | 5.17 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr1_+_16600690 | 5.13 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr18_-_39288894 | 5.07 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr8_+_25761654 | 5.07 |
ENSDART00000137899
ENSDART00000062403 |
tmem9
|
transmembrane protein 9 |
| chr23_+_19590598 | 5.04 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr20_-_36617313 | 5.01 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr16_+_28270037 | 4.98 |
ENSDART00000059035
|
mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr11_-_7078392 | 4.93 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr19_+_21362553 | 4.86 |
ENSDART00000122002
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr21_+_11923701 | 4.85 |
ENSDART00000109292
|
ubap2a
|
ubiquitin associated protein 2a |
| chr25_+_36152215 | 4.82 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr7_+_22801465 | 4.76 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr4_+_5333988 | 4.73 |
ENSDART00000129398
ENSDART00000163850 ENSDART00000067374 ENSDART00000150780 ENSDART00000150493 ENSDART00000150306 |
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr15_+_17321218 | 4.72 |
ENSDART00000143796
|
cltcb
|
clathrin, heavy chain b (Hc) |
| chr2_-_32486080 | 4.70 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr12_+_15002757 | 4.69 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr11_+_714386 | 4.63 |
ENSDART00000167824
ENSDART00000187110 |
timp4.3
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 3 |
| chr5_+_15350954 | 4.59 |
ENSDART00000140990
ENSDART00000137287 ENSDART00000061653 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr21_-_21148623 | 4.56 |
ENSDART00000184364
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr11_+_16153207 | 4.55 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_-_6604623 | 4.53 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr3_-_1204341 | 4.49 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr19_-_20093341 | 4.43 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr15_-_16121496 | 4.42 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr9_-_43375205 | 4.41 |
ENSDART00000138436
|
znf385b
|
zinc finger protein 385B |
| chr3_-_33880951 | 4.32 |
ENSDART00000013228
|
cacna1aa
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a |
| chr5_+_32007627 | 4.29 |
ENSDART00000183061
|
scai
|
suppressor of cancer cell invasion |
| chr9_-_10532591 | 4.26 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr21_-_32467799 | 4.24 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr7_-_67842997 | 4.22 |
ENSDART00000169763
|
pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr21_+_5915041 | 4.20 |
ENSDART00000151370
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr14_+_45406299 | 4.16 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr22_+_4488454 | 4.14 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr22_-_3449282 | 4.12 |
ENSDART00000136798
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr16_+_29043813 | 4.02 |
ENSDART00000122681
|
nes
|
nestin |
| chr6_-_53282572 | 3.96 |
ENSDART00000172615
ENSDART00000165036 |
rbm5
|
RNA binding motif protein 5 |
| chr8_+_8712446 | 3.95 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr6_-_30658755 | 3.88 |
ENSDART00000065215
ENSDART00000181302 |
lurap1
|
leucine rich adaptor protein 1 |
| chr25_-_16826219 | 3.86 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr1_-_5394752 | 3.85 |
ENSDART00000103724
ENSDART00000188453 |
ndfip2
|
Nedd4 family interacting protein 2 |
| chr14_+_34971554 | 3.85 |
ENSDART00000184271
|
rnf145a
|
ring finger protein 145a |
| chr7_+_17716601 | 3.76 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chr4_+_5334439 | 3.75 |
ENSDART00000180644
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr9_-_34509997 | 3.73 |
ENSDART00000169114
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr11_-_3954691 | 3.73 |
ENSDART00000182041
|
pbrm1
|
polybromo 1 |
| chr15_-_22074315 | 3.72 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr5_-_21030934 | 3.71 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr15_+_24737599 | 3.68 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr7_+_21887307 | 3.62 |
ENSDART00000052871
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr16_+_10841163 | 3.57 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr24_+_26406770 | 3.56 |
ENSDART00000110011
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr16_-_21047483 | 3.55 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr7_+_46252993 | 3.54 |
ENSDART00000167149
|
znf536
|
zinc finger protein 536 |
| chr2_-_33687214 | 3.54 |
ENSDART00000147439
|
atp6v0b
|
ATPase H+ transporting V0 subunit b |
| chr15_-_19128705 | 3.44 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr24_-_23323526 | 3.43 |
ENSDART00000112256
ENSDART00000176903 |
zfhx4
|
zinc finger homeobox 4 |
| chr18_+_924949 | 3.42 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr10_+_33895315 | 3.42 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr6_-_19683406 | 3.40 |
ENSDART00000158041
|
cfap52
|
cilia and flagella associated protein 52 |
| chr14_-_24095414 | 3.40 |
ENSDART00000172747
ENSDART00000173146 |
si:ch211-277c7.7
|
si:ch211-277c7.7 |
| chr16_-_31790285 | 3.36 |
ENSDART00000184655
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr1_-_45039726 | 3.27 |
ENSDART00000186188
|
smu1b
|
SMU1, DNA replication regulator and spliceosomal factor b |
| chr7_-_26457208 | 3.24 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr9_-_34500197 | 3.24 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr15_-_28480546 | 3.15 |
ENSDART00000057696
ENSDART00000160858 |
git1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr2_-_29485408 | 3.12 |
ENSDART00000013411
|
cahz
|
carbonic anhydrase |
| chr21_-_32467099 | 3.10 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr21_-_39566854 | 3.03 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr17_-_8268406 | 2.99 |
ENSDART00000149873
ENSDART00000064668 ENSDART00000148403 |
ahi1
|
Abelson helper integration site 1 |
| chr16_+_22865942 | 2.97 |
ENSDART00000103235
ENSDART00000143957 |
flad1
|
flavin adenine dinucleotide synthetase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfil3-5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 30.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 3.7 | 14.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 2.5 | 32.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 2.3 | 9.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 2.0 | 6.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.8 | 9.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 1.7 | 5.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 1.6 | 17.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 1.5 | 21.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.3 | 6.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 1.2 | 8.4 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 1.2 | 11.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.2 | 10.5 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 1.1 | 8.0 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.1 | 19.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 1.0 | 4.0 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 1.0 | 3.0 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 1.0 | 17.8 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 1.0 | 5.9 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.9 | 11.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.9 | 2.7 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.9 | 20.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.8 | 4.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.8 | 10.7 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.8 | 4.7 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.8 | 2.4 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.8 | 3.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.8 | 4.6 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.7 | 33.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.7 | 16.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.7 | 7.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.7 | 9.6 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.7 | 2.0 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.7 | 7.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.7 | 2.0 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.7 | 13.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.6 | 4.5 | GO:0036268 | swimming(GO:0036268) |
| 0.6 | 2.6 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.6 | 12.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.6 | 2.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.6 | 2.8 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.6 | 1.7 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.5 | 5.3 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.5 | 2.6 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.5 | 2.0 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.5 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.5 | 6.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.5 | 8.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.5 | 10.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.5 | 4.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.4 | 2.6 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.4 | 1.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.4 | 2.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.4 | 2.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.4 | 5.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.4 | 3.9 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.3 | 2.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.3 | 1.0 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.3 | 9.8 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.3 | 1.0 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.3 | 5.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 20.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.3 | 2.8 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.3 | 6.0 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.3 | 19.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.3 | 0.9 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.3 | 2.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 2.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) pronephric proximal tubule development(GO:0035776) |
| 0.3 | 23.4 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.3 | 1.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 31.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.3 | 9.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.3 | 8.7 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 0.8 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 8.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.3 | 3.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.3 | 5.3 | GO:0048667 | cell morphogenesis involved in neuron differentiation(GO:0048667) |
| 0.3 | 1.8 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 0.9 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.2 | 2.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 14.5 | GO:0003146 | heart jogging(GO:0003146) |
| 0.2 | 2.2 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 4.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.8 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) dGDP metabolic process(GO:0046066) |
| 0.2 | 1.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.2 | 1.6 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 2.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 1.9 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.2 | 7.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 6.3 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.2 | 0.9 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 0.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 2.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 3.6 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 3.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 1.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 40.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.2 | 1.6 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 | 4.8 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.2 | 6.3 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.2 | 11.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 1.7 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.2 | 28.5 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.2 | 3.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 4.8 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 1.0 | GO:0009092 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.1 | 2.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 0.7 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 4.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 2.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.7 | GO:0031643 | positive regulation of myelination(GO:0031643) positive regulation of neurological system process(GO:0031646) |
| 0.1 | 5.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 4.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 12.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 0.2 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 4.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 9.2 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.1 | 0.5 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 1.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 5.0 | GO:0006757 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.1 | 3.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 1.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 10.0 | GO:0021782 | glial cell development(GO:0021782) |
| 0.1 | 1.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 6.5 | GO:0001933 | negative regulation of protein phosphorylation(GO:0001933) |
| 0.1 | 1.8 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 1.7 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 3.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 10.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.9 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 6.1 | GO:0048066 | developmental pigmentation(GO:0048066) |
| 0.1 | 2.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 20.5 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.1 | 8.9 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.1 | 25.9 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.1 | 4.0 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 10.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 4.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 1.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 5.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 2.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 1.0 | GO:0072175 | epithelial tube formation(GO:0072175) |
| 0.0 | 0.6 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 7.1 | GO:0099537 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) trans-synaptic signaling(GO:0099537) |
| 0.0 | 1.6 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 3.6 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 9.8 | GO:0001944 | vasculature development(GO:0001944) |
| 0.0 | 6.5 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 1.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.6 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 3.9 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 1.7 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.9 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 2.2 | 22.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 2.0 | 30.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.6 | 7.8 | GO:0005883 | neurofilament(GO:0005883) |
| 1.0 | 6.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 1.0 | 12.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.9 | 4.7 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.9 | 29.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.8 | 11.0 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.8 | 4.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.8 | 6.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.6 | 12.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.6 | 2.5 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.6 | 18.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.6 | 36.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.5 | 8.4 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.5 | 9.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.5 | 4.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.5 | 3.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.5 | 3.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.4 | 7.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.4 | 5.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 2.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.4 | 8.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.4 | 60.6 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.4 | 1.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 21.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.4 | 6.8 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 14.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.3 | 20.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 10.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 17.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 30.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.3 | 3.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 10.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 2.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 4.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 3.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 7.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 2.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 3.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 16.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 2.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 3.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 4.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 8.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.0 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 55.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 16.9 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 1.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 5.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 2.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.4 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 4.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 5.6 | GO:0099568 | cytoplasmic region(GO:0099568) |
| 0.0 | 3.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 12.3 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 8.9 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 19.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 2.6 | 10.5 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 2.3 | 30.5 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 2.0 | 10.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 1.9 | 5.7 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 1.7 | 24.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 1.5 | 70.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 1.4 | 5.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.3 | 9.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 1.0 | 4.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 1.0 | 3.0 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 1.0 | 4.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.9 | 13.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.9 | 7.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.9 | 13.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.8 | 5.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.8 | 21.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.8 | 18.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.7 | 7.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.7 | 2.9 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.7 | 15.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.7 | 8.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.7 | 16.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.7 | 4.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.7 | 14.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.6 | 2.6 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.6 | 2.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.6 | 30.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.6 | 14.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.6 | 3.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 2.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.5 | 2.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 5.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 20.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.5 | 4.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.4 | 5.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.4 | 6.0 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.4 | 1.6 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.4 | 4.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 5.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 2.7 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.3 | 4.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 1.2 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.3 | 2.4 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.3 | 9.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 2.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 3.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 1.0 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 4.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 0.9 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 1.8 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 2.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 2.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.1 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 2.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 23.6 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.2 | 6.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 5.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 2.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 9.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 2.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 0.5 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 1.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 6.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 7.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 48.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.5 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 49.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 1.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 3.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 5.1 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 4.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 2.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 12.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 23.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 11.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.8 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 2.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 2.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 11.0 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.1 | 4.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 3.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.7 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 4.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 17.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 1.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 8.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.4 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.7 | GO:0016896 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 1.9 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 4.3 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 35.9 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.8 | 7.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.7 | 20.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.5 | 3.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.5 | 10.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 8.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.2 | 4.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 4.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.7 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 1.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 2.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.1 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 1.4 | 23.4 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.9 | 10.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.5 | 3.7 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.5 | 12.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.5 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.5 | 10.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 7.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 6.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 5.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.3 | 2.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 3.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.3 | 7.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 3.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 2.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 2.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 2.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 9.4 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 5.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 3.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.5 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 4.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.6 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |