Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
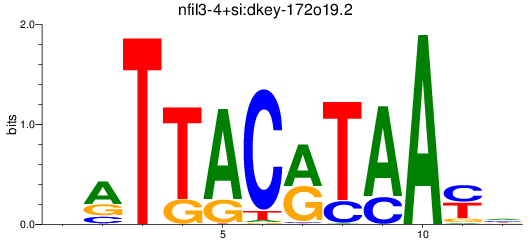
Results for nfil3-4+si:dkey-172o19.2
Z-value: 1.17
Transcription factors associated with nfil3-4+si:dkey-172o19.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-172o19.2
|
ENSDARG00000071398 | si_dkey-172o19.2 |
|
nfil3-4
|
ENSDARG00000092346 | nuclear factor, interleukin 3 regulated, member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-172o19.2 | dr11_v1_chr22_+_20546612_20546612 | 0.16 | 1.1e-01 | Click! |
| nfil3-4 | dr11_v1_chr22_+_20560041_20560041 | 0.14 | 1.9e-01 | Click! |
Activity profile of nfil3-4+si:dkey-172o19.2 motif
Sorted Z-values of nfil3-4+si:dkey-172o19.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_26310854 | 9.50 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr4_-_12725513 | 8.27 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr25_-_13188678 | 7.26 |
ENSDART00000125754
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr5_+_29831235 | 7.25 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr1_-_10071422 | 6.35 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr9_-_45602978 | 6.18 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr11_+_25278772 | 6.12 |
ENSDART00000188630
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr14_+_21107032 | 6.12 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr3_+_49043917 | 5.96 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr18_-_5598958 | 5.91 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr14_+_21106444 | 5.85 |
ENSDART00000075744
ENSDART00000132363 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr24_-_26328721 | 5.78 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr1_+_14253118 | 5.63 |
ENSDART00000161996
|
cxcl8a
|
chemokine (C-X-C motif) ligand 8a |
| chr25_-_8602437 | 5.51 |
ENSDART00000171200
|
rhcgb
|
Rh family, C glycoprotein b |
| chr11_+_43419809 | 5.46 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr18_-_7143920 | 5.25 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr17_+_26965351 | 5.12 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr6_+_42475730 | 5.12 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr5_-_30615901 | 5.08 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr14_+_3507326 | 5.06 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr8_-_36287046 | 5.03 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr11_-_6265574 | 4.96 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr7_-_24364536 | 4.90 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr5_+_4332220 | 4.87 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr14_+_1170968 | 4.85 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr13_+_33688474 | 4.83 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr14_-_12071679 | 4.77 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr5_+_57658898 | 4.74 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr20_-_26551210 | 4.72 |
ENSDART00000077715
|
si:dkey-25e12.3
|
si:dkey-25e12.3 |
| chr14_+_48862987 | 4.69 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr14_-_12071447 | 4.68 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr7_-_3429874 | 4.67 |
ENSDART00000132330
|
si:ch211-285c6.1
|
si:ch211-285c6.1 |
| chr5_+_45677781 | 4.65 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr14_+_4151379 | 4.56 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr22_+_10606573 | 4.55 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr19_+_5674907 | 4.52 |
ENSDART00000042189
|
pdk2b
|
pyruvate dehydrogenase kinase, isozyme 2b |
| chr18_+_13162728 | 4.52 |
ENSDART00000101472
|
tat
|
tyrosine aminotransferase |
| chr15_-_29348212 | 4.49 |
ENSDART00000133117
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr22_+_10606863 | 4.43 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr16_-_19568388 | 4.35 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr22_+_5478353 | 4.28 |
ENSDART00000160596
|
tppp
|
tubulin polymerization promoting protein |
| chr13_-_25750910 | 4.25 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr2_-_20923864 | 4.23 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr8_-_39739627 | 4.22 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr14_+_11430796 | 4.19 |
ENSDART00000165275
|
si:ch211-153b23.3
|
si:ch211-153b23.3 |
| chr18_-_7539166 | 4.15 |
ENSDART00000133541
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr25_+_13191391 | 4.15 |
ENSDART00000109937
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr16_+_40954481 | 4.09 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr24_+_38671054 | 4.07 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr7_+_21787507 | 4.07 |
ENSDART00000100936
|
tmem88b
|
transmembrane protein 88 b |
| chr10_-_22095505 | 4.06 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr13_-_20540790 | 4.03 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr17_-_2039511 | 3.99 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr5_-_65037525 | 3.95 |
ENSDART00000158856
|
anxa1b
|
annexin A1b |
| chr9_-_48736388 | 3.95 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr8_-_36475328 | 3.90 |
ENSDART00000048448
|
si:busm1-266f07.2
|
si:busm1-266f07.2 |
| chr3_-_16142057 | 3.89 |
ENSDART00000016616
|
arl5c
|
ADP-ribosylation factor-like 5C |
| chr24_-_2843107 | 3.87 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr7_+_30392613 | 3.85 |
ENSDART00000075508
|
lipca
|
lipase, hepatic a |
| chr11_-_8167799 | 3.85 |
ENSDART00000133574
ENSDART00000024046 ENSDART00000146940 |
uox
|
urate oxidase |
| chr5_-_65037371 | 3.84 |
ENSDART00000170560
|
anxa1b
|
annexin A1b |
| chr16_+_23984755 | 3.83 |
ENSDART00000145328
|
apoc2
|
apolipoprotein C-II |
| chr22_+_11775269 | 3.82 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr14_+_16345003 | 3.81 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr3_-_50139860 | 3.80 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr25_+_13191615 | 3.75 |
ENSDART00000168849
|
si:ch211-147m6.2
|
si:ch211-147m6.2 |
| chr9_-_16109001 | 3.73 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr11_-_23501467 | 3.69 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr8_-_21103041 | 3.68 |
ENSDART00000171771
ENSDART00000131322 ENSDART00000137838 |
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr3_+_30921246 | 3.66 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr6_+_9130989 | 3.62 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr16_+_50289916 | 3.61 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr13_-_31008275 | 3.59 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr2_-_39017838 | 3.58 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr23_-_23401305 | 3.57 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr5_-_69948099 | 3.54 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr22_+_22004082 | 3.54 |
ENSDART00000148375
|
gna15.2
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 2 |
| chr13_-_25774183 | 3.51 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr16_+_46410520 | 3.50 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr17_+_27456804 | 3.48 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr22_-_13165186 | 3.47 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr13_+_7387822 | 3.47 |
ENSDART00000148240
|
exoc3l4
|
exocyst complex component 3-like 4 |
| chr16_+_23398369 | 3.46 |
ENSDART00000037694
|
s100a10b
|
S100 calcium binding protein A10b |
| chr7_+_56577522 | 3.46 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr20_+_25563105 | 3.37 |
ENSDART00000063100
|
cyp2p6
|
cytochrome P450, family 2, subfamily P, polypeptide 6 |
| chr5_-_55560937 | 3.36 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr15_+_19838458 | 3.35 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr7_+_48761875 | 3.34 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr18_+_13164325 | 3.33 |
ENSDART00000189057
|
tat
|
tyrosine aminotransferase |
| chr12_-_9132682 | 3.33 |
ENSDART00000066471
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr24_-_23784701 | 3.32 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr12_-_20373058 | 3.31 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr5_-_26181863 | 3.27 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr22_-_15593824 | 3.25 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr16_+_23397785 | 3.22 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr11_+_25504215 | 3.22 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr3_-_53465223 | 3.20 |
ENSDART00000057123
ENSDART00000125515 ENSDART00000143096 |
nr5a5
|
nuclear receptor subfamily 5, group A, member 5 |
| chr16_+_29492749 | 3.20 |
ENSDART00000179680
|
ctsk
|
cathepsin K |
| chr16_+_21801277 | 3.18 |
ENSDART00000088407
|
trim108
|
tripartite motif containing 108 |
| chr16_+_23487051 | 3.16 |
ENSDART00000145496
|
icn2
|
ictacalcin 2 |
| chr17_-_20236228 | 3.15 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr5_-_2112030 | 3.15 |
ENSDART00000091932
|
gusb
|
glucuronidase, beta |
| chr22_-_24297510 | 3.14 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr2_+_10134345 | 3.13 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr14_+_21699129 | 3.09 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr4_-_20181964 | 3.08 |
ENSDART00000022539
|
fgl2a
|
fibrinogen-like 2a |
| chr5_+_2815021 | 3.08 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr4_-_27099224 | 3.08 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr2_+_24762567 | 3.08 |
ENSDART00000078866
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr7_+_38380135 | 3.07 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr10_-_21941443 | 3.05 |
ENSDART00000174954
|
FO744833.1
|
|
| chr8_-_21103522 | 3.04 |
ENSDART00000100283
|
aldh9a1a.1
|
aldehyde dehydrogenase 9 family, member A1a, tandem duplicate 1 |
| chr22_-_10121880 | 3.02 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr19_-_15281996 | 3.02 |
ENSDART00000103784
|
edn2
|
endothelin 2 |
| chr18_+_48802154 | 3.02 |
ENSDART00000191403
|
bmp16
|
bone morphogenetic protein 16 |
| chr2_+_26288301 | 3.00 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr13_-_36034582 | 2.99 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr20_-_30900947 | 2.99 |
ENSDART00000153419
ENSDART00000062536 |
hebp2
|
heme binding protein 2 |
| chr20_-_25533739 | 2.98 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr25_+_14398650 | 2.96 |
ENSDART00000130548
|
ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr22_+_980290 | 2.93 |
ENSDART00000065377
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr17_-_6076266 | 2.89 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr2_+_20793982 | 2.89 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr8_+_21225064 | 2.88 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr6_+_36381709 | 2.87 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr23_+_5108374 | 2.86 |
ENSDART00000114263
|
zgc:194242
|
zgc:194242 |
| chr12_-_5120339 | 2.84 |
ENSDART00000168759
|
rbp4
|
retinol binding protein 4, plasma |
| chr25_+_6122823 | 2.84 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr25_-_29415369 | 2.82 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr3_+_34670076 | 2.81 |
ENSDART00000133457
|
dlx4a
|
distal-less homeobox 4a |
| chr3_-_26017592 | 2.80 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr5_-_48680580 | 2.80 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr1_-_20928772 | 2.77 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr7_+_15736230 | 2.75 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr22_-_37686966 | 2.74 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr13_+_7049823 | 2.74 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr2_+_43920461 | 2.73 |
ENSDART00000123673
|
si:ch211-195h23.3
|
si:ch211-195h23.3 |
| chr1_-_39895859 | 2.72 |
ENSDART00000135791
ENSDART00000035739 |
tmem134
|
transmembrane protein 134 |
| chr3_+_4403980 | 2.70 |
ENSDART00000156881
|
CR774195.1
|
|
| chr5_-_30074332 | 2.69 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr20_-_25522911 | 2.68 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr7_-_19168375 | 2.67 |
ENSDART00000112447
|
il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr21_-_17956416 | 2.67 |
ENSDART00000026737
|
stx2a
|
syntaxin 2a |
| chr5_+_27421025 | 2.66 |
ENSDART00000098590
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr7_+_48761646 | 2.66 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr25_+_32473433 | 2.65 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr8_-_41228530 | 2.65 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr5_-_12587053 | 2.65 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr1_+_25696798 | 2.64 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a |
| chr1_-_18615063 | 2.63 |
ENSDART00000014916
|
klf3
|
Kruppel-like factor 3 (basic) |
| chr15_-_35960250 | 2.63 |
ENSDART00000186765
|
col4a4
|
collagen, type IV, alpha 4 |
| chr5_+_26121393 | 2.61 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr8_-_11324143 | 2.60 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr7_-_25133783 | 2.60 |
ENSDART00000173781
ENSDART00000121943 ENSDART00000077219 |
badb
|
BCL2 associated agonist of cell death b |
| chr5_+_37091626 | 2.59 |
ENSDART00000161054
|
tagln2
|
transgelin 2 |
| chr9_+_41224100 | 2.58 |
ENSDART00000141792
|
stat1b
|
signal transducer and activator of transcription 1b |
| chr11_+_20896122 | 2.58 |
ENSDART00000162339
ENSDART00000181111 |
CABZ01008730.1
|
|
| chr2_-_55337585 | 2.57 |
ENSDART00000177924
|
tpm4b
|
tropomyosin 4b |
| chr12_+_5708400 | 2.57 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr1_-_59240975 | 2.56 |
ENSDART00000166170
|
mvb12a
|
multivesicular body subunit 12A |
| chr22_+_1796057 | 2.54 |
ENSDART00000170834
|
znf1179
|
zinc finger protein 1179 |
| chr6_-_14010554 | 2.53 |
ENSDART00000004656
|
zgc:92027
|
zgc:92027 |
| chr1_-_2457546 | 2.53 |
ENSDART00000103795
|
ggact.1
|
gamma-glutamylamine cyclotransferase, tandem duplicate 1 |
| chr3_+_16922226 | 2.53 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr16_+_11834516 | 2.52 |
ENSDART00000146611
|
cxcr3.3
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 3 |
| chr15_+_41919484 | 2.48 |
ENSDART00000099821
ENSDART00000146246 |
nlrp16
|
NACHT, LRR and PYD domains-containing protein 16 |
| chr1_+_11659861 | 2.48 |
ENSDART00000054787
|
BX569798.1
|
|
| chr2_-_41861040 | 2.48 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr7_-_39552314 | 2.46 |
ENSDART00000134174
|
slc22a18
|
solute carrier family 22, member 18 |
| chr12_-_25294769 | 2.45 |
ENSDART00000153306
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr20_-_40755614 | 2.44 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr19_-_30811161 | 2.44 |
ENSDART00000103524
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr20_-_27733683 | 2.43 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr7_-_3894831 | 2.43 |
ENSDART00000172921
|
si:dkey-88n24.11
|
si:dkey-88n24.11 |
| chr7_+_19424857 | 2.42 |
ENSDART00000173674
|
si:ch211-212k18.6
|
si:ch211-212k18.6 |
| chr12_-_5120175 | 2.41 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr2_+_35595454 | 2.41 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr7_+_56577906 | 2.38 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr8_-_30791266 | 2.38 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr17_+_45648836 | 2.37 |
ENSDART00000155037
|
zgc:162184
|
zgc:162184 |
| chr7_+_39706004 | 2.36 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr16_+_23984179 | 2.36 |
ENSDART00000175879
|
apoc2
|
apolipoprotein C-II |
| chr25_-_26833100 | 2.35 |
ENSDART00000014052
|
neil1
|
nei-like DNA glycosylase 1 |
| chr11_+_37265692 | 2.34 |
ENSDART00000184691
|
il17rc
|
interleukin 17 receptor C |
| chr3_-_26017831 | 2.34 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr19_+_14113886 | 2.33 |
ENSDART00000169343
|
kdf1b
|
keratinocyte differentiation factor 1b |
| chr18_+_12655766 | 2.33 |
ENSDART00000144246
|
tbxas1
|
thromboxane A synthase 1 (platelet) |
| chr19_-_977849 | 2.32 |
ENSDART00000172303
|
CABZ01088282.1
|
|
| chr21_-_13668358 | 2.31 |
ENSDART00000180323
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr22_-_26175237 | 2.31 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr2_-_127945 | 2.30 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr20_-_9760424 | 2.30 |
ENSDART00000104936
|
si:dkey-63j12.4
|
si:dkey-63j12.4 |
| chr7_+_3597413 | 2.29 |
ENSDART00000064281
|
si:dkey-192d15.2
|
si:dkey-192d15.2 |
| chr3_-_50147160 | 2.28 |
ENSDART00000191341
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr16_+_46725087 | 2.28 |
ENSDART00000008920
|
rab11al
|
RAB11a, member RAS oncogene family, like |
| chr2_+_17055069 | 2.27 |
ENSDART00000115078
|
thpo
|
thrombopoietin |
| chr16_+_32029090 | 2.27 |
ENSDART00000041054
|
tmc4
|
transmembrane channel-like 4 |
| chr23_+_19814371 | 2.26 |
ENSDART00000182897
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr18_+_2228737 | 2.25 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr25_+_24616717 | 2.25 |
ENSDART00000089113
|
abtb2b
|
ankyrin repeat and BTB (POZ) domain containing 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfil3-4+si:dkey-172o19.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 1.8 | 10.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.7 | 5.1 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 1.7 | 5.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 1.6 | 4.7 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 1.5 | 4.4 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 1.3 | 7.8 | GO:0033028 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 1.3 | 3.8 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 1.3 | 8.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.3 | 3.8 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 1.2 | 4.9 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 1.2 | 12.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 1.2 | 9.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.1 | 4.5 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 1.1 | 3.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 1.1 | 4.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.0 | 6.0 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 1.0 | 2.9 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 1.0 | 3.9 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 1.0 | 7.6 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.9 | 1.9 | GO:2000108 | positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.9 | 2.6 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.9 | 2.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.8 | 4.0 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.8 | 0.8 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.8 | 3.8 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.8 | 3.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.8 | 5.3 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.8 | 3.0 | GO:0060471 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.7 | 4.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.7 | 2.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.7 | 2.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.7 | 10.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.7 | 2.0 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.6 | 1.9 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.6 | 1.9 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.6 | 8.8 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.6 | 1.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 2.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.6 | 1.8 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.6 | 8.2 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.6 | 1.7 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.5 | 2.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 1.6 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.5 | 2.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.5 | 5.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.5 | 3.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.5 | 3.6 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.5 | 1.5 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.5 | 1.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.5 | 3.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.5 | 3.4 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.5 | 15.3 | GO:0007568 | aging(GO:0007568) |
| 0.5 | 2.3 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.5 | 1.8 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.4 | 3.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.4 | 6.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 5.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.4 | 2.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.4 | 1.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.4 | 2.0 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.4 | 4.0 | GO:0032262 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.4 | 39.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.4 | 1.6 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.4 | 1.5 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.4 | 3.7 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 0.3 | 3.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 5.9 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.3 | 1.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 2.0 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.3 | 1.3 | GO:0009078 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.3 | 4.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.3 | 1.6 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 5.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.3 | 8.2 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.3 | 0.6 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.3 | 3.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.3 | 1.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 1.2 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.3 | 0.3 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.3 | 0.9 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.3 | 4.5 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.3 | 0.8 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.3 | 10.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 4.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 6.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 5.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 0.8 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.3 | 1.0 | GO:0099625 | regulation of membrane repolarization(GO:0060306) regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.3 | 2.6 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.3 | 1.5 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 2.7 | GO:0042493 | response to drug(GO:0042493) |
| 0.2 | 1.7 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.2 | 1.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 1.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 4.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 1.0 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.2 | 2.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 0.5 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 2.9 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.2 | 0.9 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.2 | 4.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.2 | 1.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.6 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 | 1.1 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.2 | 1.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.2 | 1.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 2.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 2.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 2.5 | GO:0045851 | phagolysosome assembly(GO:0001845) vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 2.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.2 | 1.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.9 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.2 | 1.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 3.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 0.6 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.2 | 0.9 | GO:0046461 | neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.2 | 1.8 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.2 | 2.0 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 1.1 | GO:0031282 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.2 | 2.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 0.9 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.2 | 1.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 0.5 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 1.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 1.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 3.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.2 | 4.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 1.0 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 5.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 1.3 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.2 | 0.9 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 6.1 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.2 | 1.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 3.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.2 | 0.6 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 2.7 | GO:0009749 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.2 | 0.6 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 3.0 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.9 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 1.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 5.8 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.9 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 0.6 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 2.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.6 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.1 | 2.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 1.2 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.5 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.4 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 4.6 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 3.1 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.1 | 0.5 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.5 | GO:0019477 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 2.6 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 7.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 1.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.5 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.4 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.5 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 2.1 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.1 | 0.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 3.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.0 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.6 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 1.1 | GO:0010884 | positive regulation of lipid storage(GO:0010884) regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.1 | 0.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 2.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 3.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 3.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 1.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.6 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 1.1 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.9 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 1.3 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.9 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.8 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.8 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.1 | 2.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 2.4 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 2.1 | GO:0044275 | cellular carbohydrate catabolic process(GO:0044275) |
| 0.1 | 0.5 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 1.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 1.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.1 | GO:0030810 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of cyclase activity(GO:0031281) positive regulation of adenylate cyclase activity(GO:0045762) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.1 | 0.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.4 | GO:0046503 | glycerophospholipid catabolic process(GO:0046475) glycerolipid catabolic process(GO:0046503) |
| 0.1 | 1.1 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 1.1 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.1 | 0.9 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 2.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.2 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 7.3 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 3.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 1.3 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.1 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.0 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.1 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.7 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.1 | 0.4 | GO:0035627 | intracellular lipid transport(GO:0032365) ceramide transport(GO:0035627) |
| 0.1 | 2.1 | GO:1902652 | cholesterol metabolic process(GO:0008203) secondary alcohol metabolic process(GO:1902652) |
| 0.1 | 1.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.3 | GO:0032979 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 3.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 2.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 2.0 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 1.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.7 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.1 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 2.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 3.4 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 5.1 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 3.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 0.2 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 3.6 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.1 | 2.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 2.2 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.1 | 0.7 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 1.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.0 | GO:0098659 | inorganic cation import into cell(GO:0098659) inorganic ion import into cell(GO:0099587) |
| 0.0 | 4.7 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) trehalose metabolic process(GO:0005991) |
| 0.0 | 5.4 | GO:0034097 | response to cytokine(GO:0034097) |
| 0.0 | 1.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.7 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.7 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.4 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.9 | GO:0015671 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.8 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 1.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.3 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.0 | 2.0 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 1.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 3.6 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.6 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0032616 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.4 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 2.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.9 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 2.3 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.0 | 1.2 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 1.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.6 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 1.0 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.6 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 3.2 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.2 | GO:0001816 | cytokine production(GO:0001816) |
| 0.0 | 1.2 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 2.5 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 1.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.0 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.8 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.8 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 1.3 | GO:0034249 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 0.7 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 1.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.9 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 1.8 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.8 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 6.3 | GO:0006955 | immune response(GO:0006955) |
| 0.0 | 0.8 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.8 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 0.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.7 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 4.6 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 1.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.2 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.3 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.5 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.2 | GO:0009208 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 0.0 | 0.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.3 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.1 | GO:0014005 | microglia development(GO:0014005) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 7.5 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.6 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.0 | 0.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.0 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.3 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.9 | GO:0009611 | response to wounding(GO:0009611) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.4 | 4.2 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 1.1 | 3.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.8 | 11.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 2.0 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.5 | 1.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.5 | 1.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.4 | 1.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 6.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.4 | 1.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.4 | 1.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 2.5 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 1.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 1.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.3 | 0.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 5.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 3.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 5.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.9 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.2 | 0.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 2.2 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.2 | 0.8 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 1.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 0.5 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.2 | 2.4 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 2.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 3.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 2.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 2.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.6 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 2.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 2.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 14.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 3.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.7 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.3 | GO:1990077 | primosome complex(GO:1990077) |
| 0.1 | 5.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 8.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 0.3 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 10.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 2.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.7 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.1 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 3.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 3.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 1.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 3.6 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.1 | 0.9 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 15.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 65.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 3.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 1.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 1.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 2.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 4.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 26.1 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 4.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.5 | GO:0030880 | RNA polymerase complex(GO:0030880) |
| 0.0 | 1.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 5.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 24.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 2.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.8 | GO:0031984 | trans-Golgi network(GO:0005802) organelle subcompartment(GO:0031984) Golgi subcompartment(GO:0098791) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 5.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.2 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 7.0 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 1.7 | 5.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 1.6 | 4.7 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 1.6 | 4.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.4 | 5.6 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 1.4 | 4.2 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 1.3 | 7.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.3 | 3.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.1 | 8.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.1 | 4.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 1.1 | 4.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.0 | 5.9 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.9 | 3.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.8 | 3.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.8 | 3.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.8 | 5.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.7 | 5.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.7 | 2.2 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.7 | 2.9 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.7 | 2.1 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.7 | 4.9 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.7 | 2.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.7 | 4.7 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.6 | 12.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.6 | 3.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.6 | 1.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 1.8 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.6 | 1.7 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.6 | 17.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.6 | 2.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.6 | 1.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.6 | 1.7 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.5 | 2.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.5 | 4.9 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.5 | 2.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.5 | 4.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.5 | 3.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 1.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 3.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 1.5 | GO:0010852 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.5 | 3.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 1.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.5 | 4.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.5 | 2.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.4 | 3.5 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 1.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.4 | 1.3 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.4 | 1.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.4 | 1.2 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.4 | 9.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.4 | 1.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.4 | 1.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.4 | 1.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 3.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.3 | 5.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 1.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.3 | 1.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 4.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 9.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.3 | 2.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.3 | 2.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 2.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.3 | 1.9 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.3 | 1.4 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 1.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.3 | 0.8 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.3 | 2.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 2.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 1.0 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.2 | 14.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 6.6 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.2 | 1.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 0.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 1.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 4.0 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 2.3 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 1.4 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.2 | 2.7 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.2 | 0.9 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.2 | 5.0 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 1.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 10.6 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.4 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 1.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.2 | 1.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 2.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 3.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 0.7 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 0.8 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.2 | 6.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.2 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.8 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.2 | 0.7 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.2 | 2.1 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 2.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 1.4 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.2 | 2.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.2 | 2.5 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 1.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 0.6 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 7.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 0.9 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.3 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 2.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 4.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 4.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 2.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.2 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.4 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 2.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 3.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 3.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.8 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 2.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 3.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 5.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 4.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.8 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 1.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.7 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 7.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.7 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 2.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 2.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.0 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 2.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 3.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.3 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.3 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 8.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 2.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 3.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 4.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.2 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.4 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 5.3 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 2.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 2.4 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 1.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 9.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 0.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 4.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 33.8 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 5.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 8.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.9 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.1 | 0.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.5 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 4.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 1.3 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 1.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 3.4 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 2.3 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 3.5 | GO:0046906 | tetrapyrrole binding(GO:0046906) |
| 0.0 | 9.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 2.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 5.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 4.7 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 3.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0050780 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 1.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 14.4 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 1.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.0 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 3.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 4.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.2 | GO:0016829 | lyase activity(GO:0016829) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.5 | 21.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 7.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 2.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 3.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 1.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 3.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 4.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 5.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 2.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 3.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 5.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.7 | 5.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.6 | 3.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 6.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.6 | 12.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.5 | 6.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.5 | 6.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.5 | 2.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.4 | 5.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 3.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 0.8 | REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.4 | 3.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 3.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 4.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 7.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.3 | 1.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 3.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 1.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.3 | 7.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 1.7 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 1.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 1.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 6.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 0.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 2.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 3.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 1.7 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 3.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 3.5 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 5.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.9 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.0 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 4.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 3.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.1 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.9 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.4 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 2.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.5 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |