Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
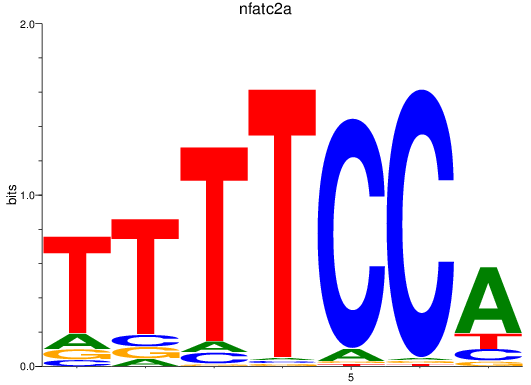
Results for nfatc2a
Z-value: 1.26
Transcription factors associated with nfatc2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc2a
|
ENSDARG00000100927 | nuclear factor of activated T cells 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfatc2a | dr11_v1_chr23_+_39089574_39089574 | -0.07 | 4.8e-01 | Click! |
Activity profile of nfatc2a motif
Sorted Z-values of nfatc2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_60551133 | 13.42 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr6_+_3827751 | 11.41 |
ENSDART00000003008
ENSDART00000122348 |
gad1b
|
glutamate decarboxylase 1b |
| chr16_-_16182319 | 10.85 |
ENSDART00000103815
|
stmn2a
|
stathmin 2a |
| chr19_-_31522625 | 10.06 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr5_-_13835461 | 9.48 |
ENSDART00000148297
ENSDART00000114841 |
add2
|
adducin 2 (beta) |
| chr6_+_48618512 | 9.36 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr4_-_15420452 | 8.73 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr22_-_13851297 | 8.71 |
ENSDART00000080306
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr8_+_22931427 | 8.61 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr6_+_13920479 | 8.40 |
ENSDART00000155480
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr8_+_26859639 | 8.16 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr18_-_38087875 | 8.04 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr2_-_13216269 | 7.99 |
ENSDART00000149947
|
bcl2b
|
BCL2, apoptosis regulator b |
| chr22_+_24715282 | 7.96 |
ENSDART00000088027
ENSDART00000189054 ENSDART00000140430 |
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr17_-_6730247 | 7.93 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr24_-_24163201 | 7.93 |
ENSDART00000140170
|
map7d2b
|
MAP7 domain containing 2b |
| chr18_+_17663898 | 7.84 |
ENSDART00000021213
|
cpne2
|
copine II |
| chr17_+_23300827 | 7.77 |
ENSDART00000058745
|
zgc:165461
|
zgc:165461 |
| chr11_-_3552067 | 7.54 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr18_-_10298162 | 7.43 |
ENSDART00000007520
|
lrrc4.2
|
leucine rich repeat containing 4.2 |
| chr1_-_56223913 | 7.42 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr23_+_37323962 | 7.29 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr9_+_29585943 | 7.24 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr2_-_36925561 | 7.13 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr8_-_7502166 | 7.07 |
ENSDART00000176938
|
cdk20
|
cyclin-dependent kinase 20 |
| chr6_-_13187168 | 7.06 |
ENSDART00000193286
ENSDART00000188350 ENSDART00000150036 ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr1_+_11691722 | 7.06 |
ENSDART00000134531
|
eif3bb
|
eukaryotic translation initiation factor 3, subunit Bb |
| chr17_-_17130942 | 7.03 |
ENSDART00000064241
|
nrxn3a
|
neurexin 3a |
| chr2_+_24199276 | 7.01 |
ENSDART00000140575
|
map4l
|
microtubule associated protein 4 like |
| chr21_-_21089781 | 7.01 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr2_-_44282796 | 7.00 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr20_+_45741566 | 6.99 |
ENSDART00000113454
|
chgb
|
chromogranin B |
| chr24_-_21921262 | 6.60 |
ENSDART00000186061
ENSDART00000187846 |
tagln3b
|
transgelin 3b |
| chr22_-_11493236 | 6.54 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr10_+_21576909 | 6.42 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr10_+_21559605 | 6.31 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr6_+_29923593 | 6.24 |
ENSDART00000169687
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr7_-_56793739 | 6.23 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr12_+_24344611 | 6.19 |
ENSDART00000093094
|
nrxn1a
|
neurexin 1a |
| chr4_+_26628822 | 6.14 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr3_-_50277959 | 6.13 |
ENSDART00000082773
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr24_-_5786759 | 6.13 |
ENSDART00000152069
|
chst2b
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2b |
| chr10_-_27049170 | 6.06 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr1_-_12278522 | 6.02 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr4_+_19535946 | 6.02 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr24_-_24162930 | 5.97 |
ENSDART00000080602
|
map7d2b
|
MAP7 domain containing 2b |
| chr8_-_14049404 | 5.97 |
ENSDART00000093117
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr5_-_29643930 | 5.90 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr7_+_15871156 | 5.90 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr14_-_33872616 | 5.83 |
ENSDART00000162840
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr7_+_30787903 | 5.81 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr25_+_6306885 | 5.78 |
ENSDART00000142705
ENSDART00000067510 |
crabp1a
|
cellular retinoic acid binding protein 1a |
| chr10_+_21701568 | 5.78 |
ENSDART00000090748
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr24_-_33756003 | 5.74 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr6_+_27667359 | 5.72 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr25_+_4837915 | 5.72 |
ENSDART00000168016
|
gnb5a
|
guanine nucleotide binding protein (G protein), beta 5a |
| chr6_-_894006 | 5.69 |
ENSDART00000171091
|
zeb2b
|
zinc finger E-box binding homeobox 2b |
| chr5_+_63668735 | 5.67 |
ENSDART00000134261
ENSDART00000097330 |
dnm1b
|
dynamin 1b |
| chr25_-_8030113 | 5.59 |
ENSDART00000104674
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr2_+_26237322 | 5.57 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr11_+_14147913 | 5.56 |
ENSDART00000022823
ENSDART00000154329 |
plppr3b
|
phospholipid phosphatase related 3b |
| chr9_+_42063906 | 5.55 |
ENSDART00000048893
|
pcbp3
|
poly(rC) binding protein 3 |
| chr9_+_44430705 | 5.54 |
ENSDART00000190696
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr15_+_29085955 | 5.54 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr25_-_22639133 | 5.54 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr25_-_19068557 | 5.53 |
ENSDART00000184780
|
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr23_+_40460333 | 5.51 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr16_-_29334672 | 5.47 |
ENSDART00000162835
|
bcan
|
brevican |
| chr10_+_21660447 | 5.38 |
ENSDART00000164519
|
pcdh1g3
|
protocadherin 1 gamma 3 |
| chr20_+_27393668 | 5.36 |
ENSDART00000005473
|
tmem179
|
transmembrane protein 179 |
| chr15_-_16098531 | 5.36 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr14_-_46616487 | 5.36 |
ENSDART00000105417
ENSDART00000166550 ENSDART00000105418 |
prom1a
|
prominin 1a |
| chr25_-_15045338 | 5.35 |
ENSDART00000161165
ENSDART00000165774 ENSDART00000172538 |
pax6a
|
paired box 6a |
| chr3_-_18274691 | 5.30 |
ENSDART00000161140
|
BX649434.3
|
|
| chr24_+_15897717 | 5.29 |
ENSDART00000105956
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr10_-_22057001 | 5.27 |
ENSDART00000016575
|
tlx3b
|
T cell leukemia homeobox 3b |
| chr5_+_52625975 | 5.22 |
ENSDART00000170341
ENSDART00000168317 |
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr8_-_14052349 | 5.21 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr13_+_28819768 | 5.21 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr12_+_7445595 | 5.20 |
ENSDART00000103536
ENSDART00000152524 |
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr9_-_27442339 | 5.19 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr6_-_52428826 | 5.18 |
ENSDART00000047399
|
mmp24
|
matrix metallopeptidase 24 |
| chr17_+_52822422 | 5.16 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr11_-_29623380 | 5.09 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr10_+_21656654 | 5.09 |
ENSDART00000160464
|
pcdh1g2
|
protocadherin 1 gamma 2 |
| chr17_-_15528597 | 5.03 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr21_+_31150773 | 5.00 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr20_-_28800999 | 4.97 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr4_-_7212875 | 4.97 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr10_+_21677058 | 4.97 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr7_-_24699985 | 4.94 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr14_-_39074539 | 4.92 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr25_-_28384954 | 4.92 |
ENSDART00000073500
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr25_-_19443421 | 4.90 |
ENSDART00000067362
|
cart2
|
cocaine- and amphetamine-regulated transcript 2 |
| chr15_-_25392589 | 4.89 |
ENSDART00000124205
|
si:dkey-54n8.4
|
si:dkey-54n8.4 |
| chr22_+_5574952 | 4.89 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr16_+_45746549 | 4.88 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr13_+_24834199 | 4.83 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr7_-_57933736 | 4.81 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr20_-_10120442 | 4.78 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr6_-_12314475 | 4.75 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr1_-_42778510 | 4.72 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr9_-_35069645 | 4.67 |
ENSDART00000122679
ENSDART00000077908 ENSDART00000077894 ENSDART00000125536 |
appb
|
amyloid beta (A4) precursor protein b |
| chr20_-_39273987 | 4.61 |
ENSDART00000127173
|
clu
|
clusterin |
| chr17_-_7371564 | 4.56 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr16_+_25316973 | 4.56 |
ENSDART00000086409
|
dync1i1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr3_+_24482999 | 4.55 |
ENSDART00000059179
|
nptxra
|
neuronal pentraxin receptor a |
| chr10_+_21786656 | 4.52 |
ENSDART00000185851
ENSDART00000167219 |
pcdh1g26
|
protocadherin 1 gamma 26 |
| chr2_-_11512819 | 4.50 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr8_-_31919624 | 4.49 |
ENSDART00000085573
|
rgs7bpa
|
regulator of G protein signaling 7 binding protein a |
| chr15_-_27710513 | 4.48 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr9_+_24159280 | 4.47 |
ENSDART00000184624
ENSDART00000178422 |
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr23_-_24450686 | 4.45 |
ENSDART00000189161
|
spen
|
spen family transcriptional repressor |
| chr20_-_32112818 | 4.41 |
ENSDART00000142653
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr20_+_10166297 | 4.41 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr5_+_37966505 | 4.40 |
ENSDART00000127648
|
pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr24_-_15648636 | 4.38 |
ENSDART00000136200
|
cbln2b
|
cerebellin 2b precursor |
| chr23_-_29505463 | 4.33 |
ENSDART00000050915
|
kif1b
|
kinesin family member 1B |
| chr5_-_69041102 | 4.31 |
ENSDART00000161561
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr12_-_3453589 | 4.27 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr20_+_18661624 | 4.27 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr4_+_11140197 | 4.27 |
ENSDART00000067264
|
cracr2ab
|
calcium release activated channel regulator 2Ab |
| chr8_+_7144066 | 4.26 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr11_-_32723851 | 4.23 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr16_+_34160835 | 4.23 |
ENSDART00000054037
|
oprd1b
|
opioid receptor, delta 1b |
| chr17_-_12336987 | 4.22 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr19_+_29798064 | 4.18 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr15_-_5467477 | 4.18 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr19_-_10243148 | 4.17 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr7_-_40993456 | 4.17 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr21_+_11468934 | 4.15 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr22_-_21046654 | 4.14 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr7_-_52842007 | 4.14 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr25_-_5740334 | 4.14 |
ENSDART00000169622
ENSDART00000168720 |
LO017739.1
|
|
| chr20_+_18209895 | 4.14 |
ENSDART00000111063
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr25_+_35250976 | 4.13 |
ENSDART00000003494
|
slc17a6a
|
solute carrier family 17 (vesicular glutamate transporter), member 6a |
| chr22_-_33679277 | 4.12 |
ENSDART00000169948
|
FO904977.1
|
|
| chr5_-_32323136 | 4.11 |
ENSDART00000110804
|
hspb15
|
heat shock protein, alpha-crystallin-related, b15 |
| chr24_-_15648878 | 4.09 |
ENSDART00000181854
|
cbln2b
|
cerebellin 2b precursor |
| chr14_-_18671334 | 4.09 |
ENSDART00000182381
|
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr19_+_6938289 | 4.09 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr9_+_22632126 | 4.06 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr3_+_15550522 | 4.06 |
ENSDART00000136912
ENSDART00000176218 |
si:dkey-93n13.3
|
si:dkey-93n13.3 |
| chr18_+_1703984 | 4.05 |
ENSDART00000114010
|
slitrk3a
|
SLIT and NTRK-like family, member 3a |
| chr21_-_42202792 | 4.04 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr25_+_31122806 | 4.03 |
ENSDART00000067039
|
rassf8a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8a |
| chr13_+_11440389 | 4.03 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr19_-_15335787 | 4.03 |
ENSDART00000187131
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr22_-_11729350 | 4.02 |
ENSDART00000105813
|
krt222
|
keratin 222 |
| chr12_+_5081759 | 4.00 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr1_+_25783801 | 3.99 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr25_+_36152215 | 3.99 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr17_+_15534815 | 3.99 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr23_+_16633951 | 3.99 |
ENSDART00000109537
ENSDART00000193323 |
snphb
|
syntaphilin b |
| chr16_+_43152727 | 3.98 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr22_+_5103349 | 3.96 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr6_-_49159207 | 3.95 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr13_-_46991577 | 3.94 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr22_+_9472814 | 3.94 |
ENSDART00000112125
ENSDART00000138850 |
cacna2d2b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2b |
| chr12_+_24344963 | 3.93 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr3_-_20040636 | 3.93 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr15_+_16521785 | 3.92 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr5_+_64319590 | 3.91 |
ENSDART00000192652
|
FQ377918.1
|
|
| chr20_+_34717403 | 3.90 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr25_-_21085661 | 3.89 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr10_+_42678520 | 3.89 |
ENSDART00000182496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr9_-_23217196 | 3.87 |
ENSDART00000083567
|
kif5c
|
kinesin family member 5C |
| chr13_+_32740509 | 3.84 |
ENSDART00000076423
ENSDART00000160138 |
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr17_-_20979077 | 3.84 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr20_-_18731268 | 3.84 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr2_-_30693742 | 3.84 |
ENSDART00000090292
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr4_+_3478049 | 3.83 |
ENSDART00000153944
|
grm8a
|
glutamate receptor, metabotropic 8a |
| chr7_-_34339845 | 3.83 |
ENSDART00000173816
|
madd
|
MAP-kinase activating death domain |
| chr14_+_14225048 | 3.83 |
ENSDART00000168749
|
nlgn3a
|
neuroligin 3a |
| chr9_-_48397702 | 3.82 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr25_-_12203952 | 3.79 |
ENSDART00000158204
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr10_+_21668606 | 3.78 |
ENSDART00000185751
|
CT737184.2
|
|
| chr17_-_35881841 | 3.78 |
ENSDART00000110040
|
sox11a
|
SRY (sex determining region Y)-box 11a |
| chr18_-_18849916 | 3.77 |
ENSDART00000142422
|
tgm2l
|
transglutaminase 2, like |
| chr16_+_45739193 | 3.77 |
ENSDART00000184852
ENSDART00000156851 ENSDART00000154704 |
paqr6
|
progestin and adipoQ receptor family member VI |
| chr2_+_24199073 | 3.77 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr1_+_14283692 | 3.77 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr7_-_52842605 | 3.77 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr23_+_19594608 | 3.76 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr16_-_31435020 | 3.71 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr5_+_36768674 | 3.71 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr13_-_11378355 | 3.70 |
ENSDART00000164566
|
akt3a
|
v-akt murine thymoma viral oncogene homolog 3a |
| chr12_-_4651988 | 3.70 |
ENSDART00000182836
|
si:ch211-255p10.4
|
si:ch211-255p10.4 |
| chr9_-_6661657 | 3.69 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr17_-_36936649 | 3.69 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr7_-_49594995 | 3.69 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr17_+_52822831 | 3.69 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr1_+_45217425 | 3.68 |
ENSDART00000179983
ENSDART00000074683 |
EVI5L
|
si:ch211-239f4.1 |
| chr13_-_11644806 | 3.66 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr5_-_55600689 | 3.66 |
ENSDART00000013229
|
gnaq
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr6_+_53349966 | 3.65 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr10_+_24504292 | 3.65 |
ENSDART00000090059
|
mtus2a
|
microtubule associated tumor suppressor candidate 2a |
| chr10_+_21776911 | 3.64 |
ENSDART00000163077
ENSDART00000186093 |
pcdh1g22
|
protocadherin 1 gamma 22 |
| chr20_-_31497300 | 3.63 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr5_-_40190949 | 3.63 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of nfatc2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 3.4 | 10.1 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 2.9 | 8.7 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 1.7 | 6.7 | GO:0010226 | response to lithium ion(GO:0010226) |
| 1.6 | 6.5 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 1.4 | 4.3 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 1.4 | 5.8 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.4 | 10.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 1.4 | 4.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 1.2 | 4.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 1.2 | 3.6 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 1.2 | 3.5 | GO:1990416 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.2 | 5.8 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 1.1 | 9.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.1 | 3.2 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 1.1 | 6.4 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 1.1 | 4.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 1.1 | 3.2 | GO:0042942 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 1.0 | 3.1 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 1.0 | 4.2 | GO:0060300 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 1.0 | 5.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 1.0 | 6.9 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 1.0 | 3.9 | GO:0048242 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) positive regulation of amine transport(GO:0051954) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 1.0 | 4.8 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.9 | 5.7 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.9 | 2.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.9 | 2.7 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.9 | 2.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.9 | 15.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.8 | 4.0 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.8 | 5.6 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.8 | 7.0 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.8 | 20.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.8 | 3.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.8 | 2.3 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.7 | 9.3 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.7 | 4.3 | GO:0021571 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.7 | 8.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.7 | 2.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.6 | 3.9 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.6 | 5.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.6 | 6.2 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.6 | 6.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.6 | 3.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.6 | 6.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.6 | 1.8 | GO:0033335 | anal fin development(GO:0033335) |
| 0.6 | 2.4 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.6 | 1.8 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.6 | 2.4 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.6 | 1.7 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.6 | 1.7 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.6 | 4.5 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.6 | 1.7 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.6 | 3.9 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.5 | 2.1 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.5 | 2.1 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.5 | 4.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.5 | 1.5 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.5 | 4.0 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.5 | 2.5 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.5 | 2.0 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.5 | 5.3 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.5 | 1.4 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.5 | 2.8 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.5 | 11.5 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.5 | 2.3 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.4 | 2.7 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 4.0 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.4 | 9.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.4 | 1.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.4 | 4.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.4 | 5.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 | 11.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.4 | 1.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.4 | 1.7 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.4 | 3.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.4 | 2.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.4 | 4.9 | GO:0001964 | startle response(GO:0001964) |
| 0.4 | 11.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.4 | 1.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.4 | 7.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.4 | 1.5 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.4 | 7.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.4 | 1.1 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.4 | 9.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.4 | 3.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 4.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.4 | 4.8 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.4 | 1.8 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.4 | 5.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 2.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.4 | 1.4 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.4 | 2.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.4 | 3.9 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.4 | 6.7 | GO:0099590 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.4 | 2.8 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.4 | 10.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.3 | 2.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.3 | 1.3 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.3 | 1.0 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.3 | 6.0 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.3 | 1.0 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.3 | 2.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.3 | 1.6 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.3 | 1.3 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.3 | 1.3 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.3 | 1.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.3 | 1.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 1.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.3 | 9.0 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.3 | 1.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 1.5 | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation(GO:0010770) |
| 0.3 | 2.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 0.9 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.3 | 4.0 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.3 | 9.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 4.6 | GO:1900372 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.3 | 70.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 8.0 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.3 | 4.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.3 | 1.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 3.8 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.3 | 15.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.3 | 2.1 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 7.9 | GO:0043266 | regulation of potassium ion transport(GO:0043266) |
| 0.3 | 0.8 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.3 | 1.3 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.3 | 0.8 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.3 | 5.0 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 2.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 6.0 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 2.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 2.5 | GO:1990709 | presynaptic active zone assembly(GO:1904071) presynaptic active zone organization(GO:1990709) |
| 0.2 | 2.0 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.2 | 8.6 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.2 | 1.2 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 | 2.0 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 6.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 0.7 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 2.6 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 6.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 2.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 4.9 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.2 | 2.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 10.3 | GO:0021761 | limbic system development(GO:0021761) |
| 0.2 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 3.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 2.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 3.8 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.2 | 4.7 | GO:0001508 | action potential(GO:0001508) |
| 0.2 | 1.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 1.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 1.3 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 1.3 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.2 | 1.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 1.0 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.2 | 2.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 0.8 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.2 | 1.6 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.2 | 1.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.8 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 2.0 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.2 | 1.0 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.2 | 6.3 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.2 | 1.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 1.7 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.2 | 3.1 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.2 | 3.0 | GO:0006984 | ER-nucleus signaling pathway(GO:0006984) |
| 0.2 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 3.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 8.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 2.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 1.1 | GO:0036268 | swimming(GO:0036268) |
| 0.2 | 1.3 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.2 | 0.7 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.2 | 2.9 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.2 | 2.1 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.2 | 3.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.2 | 5.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 2.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 2.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 0.7 | GO:0009097 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.2 | 0.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 2.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 5.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 1.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 0.2 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.2 | 0.5 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 1.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 1.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 3.6 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.2 | 0.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 1.4 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 1.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.2 | 1.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.2 | 0.8 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 0.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.9 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 3.0 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.2 | 5.0 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 4.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 1.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 9.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 1.4 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.4 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 1.7 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 5.2 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.1 | 1.8 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.1 | 0.7 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 5.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) regulation of neurotransmitter transport(GO:0051588) |
| 0.1 | 2.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 2.4 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.5 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 1.0 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 15.1 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.1 | 1.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 2.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 0.4 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 4.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 7.1 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.1 | 0.4 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.7 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 4.0 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.5 | GO:0070828 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) heterochromatin organization(GO:0070828) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.6 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.1 | 1.9 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 2.6 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.1 | 2.2 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.6 | GO:1902023 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 3.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 1.6 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 0.3 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 6.3 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.1 | 3.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 12.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 0.4 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 20.2 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 1.7 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.7 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.8 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 5.1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 1.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 4.5 | GO:0010970 | establishment of localization by movement along microtubule(GO:0010970) |
| 0.1 | 3.0 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 8.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.3 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 0.4 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 2.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 7.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.0 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.3 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.1 | 1.0 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 1.2 | GO:0060323 | head morphogenesis(GO:0060323) face morphogenesis(GO:0060325) |
| 0.1 | 3.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 2.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 2.3 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 7.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 0.3 | GO:0009202 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.1 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 1.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.5 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 8.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.4 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 2.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 1.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 4.1 | GO:0048675 | axon extension(GO:0048675) |
| 0.1 | 0.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 1.5 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.6 | GO:0002886 | mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) regulation of myeloid leukocyte mediated immunity(GO:0002886) regulation of mast cell activation involved in immune response(GO:0033006) leukocyte degranulation(GO:0043299) regulation of leukocyte degranulation(GO:0043300) mast cell degranulation(GO:0043303) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.8 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 0.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.5 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 1.3 | GO:1903307 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.1 | 0.4 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 4.0 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.1 | 0.3 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 3.0 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.1 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 5.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 1.7 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.3 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 1.4 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.1 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 2.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 3.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 0.4 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.1 | 9.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 24.3 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.1 | 1.0 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 2.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 1.1 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 1.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 11.4 | GO:0016311 | dephosphorylation(GO:0016311) |
| 0.0 | 0.3 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 1.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 2.4 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.7 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 9.9 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 0.3 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.5 | GO:0051205 | protein insertion into ER membrane(GO:0045048) protein insertion into membrane(GO:0051205) |
| 0.0 | 0.5 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 1.0 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.2 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.6 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 3.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 1.6 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.4 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.1 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.2 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.3 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.2 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.3 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.8 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.4 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 1.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.6 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.4 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.7 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 1.3 | 10.8 | GO:0071914 | prominosome(GO:0071914) |
| 1.0 | 4.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.9 | 13.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.8 | 9.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.7 | 4.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.6 | 13.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.6 | 1.7 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.6 | 3.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 20.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.5 | 3.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 2.0 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.5 | 4.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.5 | 4.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 3.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.5 | 4.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.4 | 3.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.4 | 1.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 7.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.4 | 1.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 15.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.4 | 6.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.4 | 5.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 3.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.4 | 3.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.4 | 6.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.3 | 2.8 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.3 | 4.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 19.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 2.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.3 | 1.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.3 | 14.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.3 | 1.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 3.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 6.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.3 | 4.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 9.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 14.1 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 1.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.3 | 2.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.3 | 2.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 2.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 11.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 3.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 3.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 3.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 15.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 64.2 | GO:0030425 | dendrite(GO:0030425) |
| 0.2 | 4.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 2.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 5.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 5.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 3.0 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.2 | 0.8 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 2.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 3.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.0 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 5.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 5.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.2 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 6.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.2 | 4.0 | GO:1990752 | microtubule end(GO:1990752) |
| 0.1 | 0.6 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.9 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 2.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 9.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 51.1 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 4.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.9 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 2.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 3.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 3.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 7.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 1.9 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.9 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 6.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 10.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 2.1 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 2.4 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.1 | 0.8 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.4 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 119.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 6.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 2.7 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.0 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 2.5 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 1.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 1.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.7 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.1 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 6.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.6 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 4.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 4.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 3.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 5.0 | GO:0005938 | cell cortex(GO:0005938) cytoplasmic region(GO:0099568) |
| 0.0 | 2.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 3.3 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 7.7 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 1.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.2 | 8.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.4 | 5.8 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 1.4 | 4.2 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 1.3 | 16.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 1.2 | 7.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 1.2 | 3.6 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.2 | 3.6 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 1.0 | 8.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 1.0 | 3.1 | GO:0004777 | succinate-semialdehyde dehydrogenase (NAD+) activity(GO:0004777) succinate-semialdehyde dehydrogenase [NAD(P)+] activity(GO:0009013) |
| 1.0 | 7.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.9 | 2.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.8 | 3.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.8 | 10.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.7 | 2.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.7 | 2.8 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.7 | 4.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.7 | 13.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.7 | 10.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.7 | 6.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.7 | 2.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.7 | 2.0 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.6 | 4.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.6 | 4.4 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.6 | 7.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.6 | 16.9 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.6 | 13.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.5 | 4.9 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.5 | 3.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.5 | 21.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 9.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.5 | 3.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.5 | 3.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.5 | 3.6 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.5 | 7.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 1.5 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.5 | 3.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 10.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.5 | 3.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 2.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 2.7 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.4 | 3.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 3.9 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.4 | 1.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.4 | 1.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 12.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.4 | 2.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.4 | 14.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 2.7 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.4 | 1.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 2.3 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.4 | 3.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.4 | 6.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.4 | 1.5 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 2.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.4 | 4.0 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.4 | 3.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 1.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 4.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 2.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 3.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 2.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 5.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 1.0 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.3 | 1.3 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.3 | 4.1 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.3 | 2.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.3 | 4.0 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.3 | 1.4 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.3 | 6.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.3 | 1.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 5.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.3 | 1.7 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.3 | 4.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.3 | 4.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.3 | 1.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.3 | 4.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 1.0 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 0.7 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.2 | 6.1 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.2 | 18.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 2.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 4.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 9.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 6.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.4 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.2 | 1.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 7.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 3.0 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 3.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 1.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 5.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 2.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 0.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 1.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 5.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 5.0 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 0.8 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 9.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 7.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 1.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 4.0 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 5.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 0.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.7 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 3.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 2.4 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 1.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 2.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 2.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 1.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 2.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 0.7 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 5.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 2.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.2 | 3.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 1.6 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 2.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 2.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 58.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.2 | 3.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 21.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.4 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 3.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.4 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.9 | GO:0015271 | A-type (transient outward) potassium channel activity(GO:0005250) outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 9.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.1 | 1.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 2.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 5.0 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 3.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 3.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.6 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 7.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 0.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.1 | GO:0015929 | beta-N-acetylhexosaminidase activity(GO:0004563) hexosaminidase activity(GO:0015929) |
| 0.1 | 1.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 2.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 23.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 2.0 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.8 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.5 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.1 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 4.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.2 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.1 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 2.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 1.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 4.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 0.8 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.0 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 3.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 2.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 3.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 6.7 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 11.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.7 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 26.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 3.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 11.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.3 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 2.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 7.1 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 0.3 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.5 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.1 | 0.5 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 2.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 8.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 17.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 2.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 3.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 2.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 4.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0042974 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 5.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 6.8 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 28.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.4 | GO:0030545 | receptor regulator activity(GO:0030545) receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 2.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.8 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 36.9 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 4.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 4.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 10.9 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.3 | 0.9 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 1.4 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 1.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 3.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 4.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 13.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 3.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 4.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 8.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 2.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 3.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 4.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 5.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.9 | 7.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.9 | 5.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.7 | 5.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.7 | 9.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 2.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.5 | 6.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.4 | 4.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.4 | 5.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 3.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 3.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 3.4 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.3 | 1.9 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 3.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.3 | 3.9 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 5.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 2.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 3.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.3 | 2.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.2 | 2.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 5.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 3.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 1.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 3.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.1 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |