Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
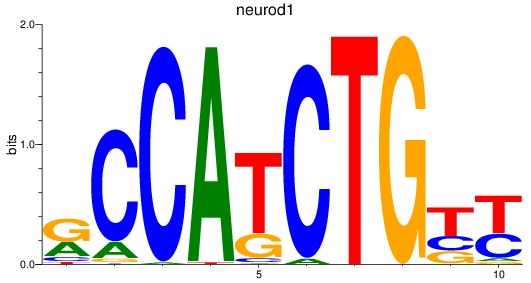
Results for neurod1
Z-value: 1.49
Transcription factors associated with neurod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
neurod1
|
ENSDARG00000019566 | neuronal differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| neurod1 | dr11_v1_chr9_-_44295071_44295071 | 0.74 | 9.3e-18 | Click! |
Activity profile of neurod1 motif
Sorted Z-values of neurod1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_29045426 | 18.70 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr8_+_7359294 | 17.50 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr9_-_44295071 | 17.35 |
ENSDART00000011837
|
neurod1
|
neuronal differentiation 1 |
| chr10_+_15777258 | 16.37 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr10_+_15777064 | 15.10 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr12_-_14922955 | 14.60 |
ENSDART00000002078
|
neurod2
|
neurogenic differentiation 2 |
| chr23_+_2778813 | 12.97 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr22_+_17205608 | 12.81 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr20_-_25669813 | 12.72 |
ENSDART00000153118
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr7_+_32722227 | 12.67 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr11_+_35364445 | 12.64 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr6_-_35487413 | 12.61 |
ENSDART00000102461
|
rgs8
|
regulator of G protein signaling 8 |
| chr19_-_32888758 | 12.23 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr23_+_20563779 | 12.16 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr13_-_30027730 | 12.13 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr16_-_29334672 | 12.11 |
ENSDART00000162835
|
bcan
|
brevican |
| chr21_+_25181003 | 11.64 |
ENSDART00000169700
|
si:dkey-183i3.9
|
si:dkey-183i3.9 |
| chr24_-_17444067 | 11.56 |
ENSDART00000155843
|
cntnap2a
|
contactin associated protein like 2a |
| chr6_-_43092175 | 11.56 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr12_-_16084835 | 11.46 |
ENSDART00000090881
|
kcnj19b
|
potassium voltage-gated channel, subfamily J, member 19b |
| chr6_-_11780070 | 11.20 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr7_+_29133321 | 11.05 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr3_-_46818001 | 11.05 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr2_+_50626476 | 10.79 |
ENSDART00000018150
|
neurod6b
|
neuronal differentiation 6b |
| chr7_+_26224211 | 10.71 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr23_-_5032587 | 10.46 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr18_-_38087875 | 10.38 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr7_+_30823749 | 10.36 |
ENSDART00000085661
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr12_-_41618844 | 10.31 |
ENSDART00000160054
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr12_+_31744217 | 10.30 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr20_+_405811 | 10.14 |
ENSDART00000149311
|
gpr63
|
G protein-coupled receptor 63 |
| chr13_+_22104298 | 10.12 |
ENSDART00000115393
|
si:dkey-174i8.1
|
si:dkey-174i8.1 |
| chr6_+_22597362 | 9.93 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr20_-_39273987 | 9.85 |
ENSDART00000127173
|
clu
|
clusterin |
| chr3_-_46817838 | 9.76 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr1_-_38709551 | 9.45 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr16_-_8927425 | 9.28 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr14_-_21219659 | 9.18 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr19_-_9882821 | 9.16 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr5_-_51819027 | 8.96 |
ENSDART00000164267
|
homer1b
|
homer scaffolding protein 1b |
| chr21_-_43952958 | 8.82 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr25_+_14017609 | 8.80 |
ENSDART00000129105
ENSDART00000125733 |
chst1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr1_-_14233815 | 8.75 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr7_-_58130703 | 8.73 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr1_-_40227166 | 8.73 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr13_+_36633355 | 8.62 |
ENSDART00000135612
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr13_+_51579851 | 8.58 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr10_-_24362775 | 8.42 |
ENSDART00000182104
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr12_-_41619257 | 8.31 |
ENSDART00000162967
|
dpysl4
|
dihydropyrimidinase-like 4 |
| chr5_-_23280098 | 8.28 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr12_+_21525496 | 8.26 |
ENSDART00000152974
|
ca10a
|
carbonic anhydrase Xa |
| chr6_+_46341306 | 8.22 |
ENSDART00000111905
|
BX649498.1
|
|
| chr5_+_3501859 | 8.17 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr5_-_10768258 | 8.09 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr25_+_7670683 | 8.07 |
ENSDART00000040275
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr5_-_40190949 | 8.06 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr16_-_29480335 | 7.91 |
ENSDART00000148930
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr23_-_18057851 | 7.89 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr18_+_48576527 | 7.78 |
ENSDART00000167555
|
kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr6_+_21095918 | 7.72 |
ENSDART00000167225
|
spega
|
SPEG complex locus a |
| chr13_-_39830999 | 7.69 |
ENSDART00000115089
|
zgc:171482
|
zgc:171482 |
| chr4_+_15968483 | 7.60 |
ENSDART00000101575
|
si:dkey-117n7.5
|
si:dkey-117n7.5 |
| chr24_+_32472155 | 7.57 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr3_+_33340939 | 7.53 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr20_-_39273505 | 7.46 |
ENSDART00000153114
|
clu
|
clusterin |
| chr18_-_38088099 | 7.44 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr21_+_13353263 | 7.42 |
ENSDART00000114677
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr3_+_50310684 | 7.31 |
ENSDART00000112152
|
gas7a
|
growth arrest-specific 7a |
| chr10_-_24371312 | 7.24 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr17_-_20717845 | 7.23 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr18_+_9635178 | 7.22 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr4_-_1324141 | 7.22 |
ENSDART00000180720
|
ptn
|
pleiotrophin |
| chr25_+_37130450 | 7.16 |
ENSDART00000183358
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr13_+_28854438 | 7.14 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr2_-_21352101 | 7.02 |
ENSDART00000057021
|
hhatla
|
hedgehog acyltransferase like, a |
| chr25_-_554142 | 6.98 |
ENSDART00000028997
|
myo9ab
|
myosin IXAb |
| chr7_-_23996133 | 6.80 |
ENSDART00000173761
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr20_-_18731268 | 6.74 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr22_-_33679277 | 6.73 |
ENSDART00000169948
|
FO904977.1
|
|
| chr14_-_32258759 | 6.69 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr21_-_16632808 | 6.59 |
ENSDART00000172645
|
unc5da
|
unc-5 netrin receptor Da |
| chr8_-_17064243 | 6.55 |
ENSDART00000185313
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr10_-_17103651 | 6.48 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr17_-_44584811 | 6.45 |
ENSDART00000165059
ENSDART00000165252 |
slc35f4
|
solute carrier family 35, member F4 |
| chr10_+_29431529 | 6.41 |
ENSDART00000158154
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr7_+_10351038 | 6.36 |
ENSDART00000173256
|
si:cabz01029535.1
|
si:cabz01029535.1 |
| chr3_-_21061931 | 6.32 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr15_+_24691088 | 6.30 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr13_+_24287093 | 6.26 |
ENSDART00000058628
|
ccsapb
|
centriole, cilia and spindle-associated protein b |
| chr25_-_16818978 | 6.19 |
ENSDART00000104140
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr10_+_17875891 | 6.13 |
ENSDART00000191744
|
phf24
|
PHD finger protein 24 |
| chr11_+_24925434 | 6.09 |
ENSDART00000131431
|
sulf2a
|
sulfatase 2a |
| chr24_+_29449690 | 6.06 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr19_-_27261102 | 6.02 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr2_-_24348948 | 5.98 |
ENSDART00000136559
|
ano8a
|
anoctamin 8a |
| chr5_-_46896541 | 5.91 |
ENSDART00000133240
|
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr11_-_28911172 | 5.89 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr5_+_36768674 | 5.87 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr13_-_40499296 | 5.82 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr1_+_32521469 | 5.77 |
ENSDART00000113818
ENSDART00000152580 |
nlgn4a
|
neuroligin 4a |
| chr18_-_39787040 | 5.74 |
ENSDART00000169916
|
dmxl2
|
Dmx-like 2 |
| chr23_+_35708730 | 5.71 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr9_+_22929675 | 5.66 |
ENSDART00000061299
|
tsn
|
translin |
| chr14_-_27121854 | 5.62 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr23_-_18057553 | 5.59 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr24_+_24461341 | 5.49 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr2_-_24348642 | 5.49 |
ENSDART00000181739
|
ano8a
|
anoctamin 8a |
| chr1_+_38775041 | 5.46 |
ENSDART00000110824
|
wdr17
|
WD repeat domain 17 |
| chr3_+_50312422 | 5.45 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr18_-_42313798 | 5.44 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr7_-_50914526 | 5.41 |
ENSDART00000160398
|
ankrd46b
|
ankyrin repeat domain 46b |
| chr21_-_12030654 | 5.37 |
ENSDART00000139145
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr12_+_5189776 | 5.35 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr11_-_32723851 | 5.35 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr17_-_36988455 | 5.30 |
ENSDART00000187180
ENSDART00000126823 |
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr19_+_24882845 | 5.26 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr21_+_7900107 | 5.25 |
ENSDART00000056560
|
ch25hl2
|
cholesterol 25-hydroxylase like 2 |
| chr4_-_9667380 | 5.23 |
ENSDART00000189671
ENSDART00000133214 |
dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr7_+_39389273 | 5.20 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr2_-_22230326 | 5.15 |
ENSDART00000127810
|
fam110b
|
family with sequence similarity 110, member B |
| chr12_-_3453589 | 5.12 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr16_-_41515662 | 5.11 |
ENSDART00000166201
ENSDART00000127243 |
siglec15l
|
sialic acid binding Ig-like lectin 15, like |
| chr6_+_54142311 | 5.10 |
ENSDART00000154115
|
hmga1b
|
high mobility group AT-hook 1b |
| chr2_+_31475772 | 5.02 |
ENSDART00000130722
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr2_+_31476065 | 4.99 |
ENSDART00000049219
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr22_+_3232925 | 4.98 |
ENSDART00000166754
|
CU929402.1
|
|
| chr13_-_44285793 | 4.96 |
ENSDART00000167383
|
CABZ01069436.1
|
|
| chr13_+_25428677 | 4.92 |
ENSDART00000186284
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr5_+_20147830 | 4.90 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr5_-_19932621 | 4.90 |
ENSDART00000088881
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr13_+_3954540 | 4.87 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr1_+_9557212 | 4.85 |
ENSDART00000111131
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr14_-_21218891 | 4.82 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr24_+_24461558 | 4.80 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr10_-_5847904 | 4.70 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr2_-_36819624 | 4.66 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr15_+_19293744 | 4.63 |
ENSDART00000184994
ENSDART00000123815 |
jam3a
|
junctional adhesion molecule 3a |
| chr11_-_37359416 | 4.58 |
ENSDART00000159184
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr6_+_58492201 | 4.57 |
ENSDART00000156375
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr2_-_9544161 | 4.49 |
ENSDART00000124425
|
slc25a24l
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24, like |
| chr9_-_4606463 | 4.46 |
ENSDART00000179110
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr24_-_34335265 | 4.46 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr8_+_36500061 | 4.45 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr16_+_11151699 | 4.44 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr16_+_46000956 | 4.42 |
ENSDART00000101753
ENSDART00000162393 |
mtmr11
|
myotubularin related protein 11 |
| chr15_+_28955004 | 4.37 |
ENSDART00000029459
|
gipr
|
gastric inhibitory polypeptide receptor |
| chr7_-_28463106 | 4.37 |
ENSDART00000137799
|
trim66
|
tripartite motif containing 66 |
| chr23_+_20705849 | 4.35 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr8_-_26609259 | 4.34 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr17_-_2535682 | 4.34 |
ENSDART00000155227
|
ccdc9b
|
coiled-coil domain containing 9B |
| chr19_+_12801940 | 4.31 |
ENSDART00000040073
|
mc5ra
|
melanocortin 5a receptor |
| chr14_-_33083539 | 4.28 |
ENSDART00000160173
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr12_+_5190049 | 4.28 |
ENSDART00000126667
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr5_-_38342992 | 4.26 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr5_+_42912966 | 4.20 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr22_+_465269 | 4.17 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr5_-_46329880 | 4.15 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr17_-_52579709 | 4.14 |
ENSDART00000156806
|
rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr1_+_21937201 | 4.11 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr20_-_44576949 | 4.10 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr3_-_60886984 | 4.09 |
ENSDART00000170974
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr6_-_10320676 | 4.08 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr4_-_20081621 | 4.04 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr21_+_39100289 | 4.01 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr23_-_18057270 | 4.01 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr12_+_26632448 | 4.00 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr13_-_8785610 | 3.99 |
ENSDART00000021083
|
calm2b
|
calmodulin 2b, (phosphorylase kinase, delta) |
| chr11_-_11625369 | 3.95 |
ENSDART00000112328
|
si:dkey-28e7.3
|
si:dkey-28e7.3 |
| chr24_+_166892 | 3.94 |
ENSDART00000100417
|
lancl2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr9_+_28693592 | 3.93 |
ENSDART00000110198
|
zgc:162780
|
zgc:162780 |
| chr5_-_40734045 | 3.93 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr19_+_33553586 | 3.90 |
ENSDART00000183477
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr8_+_8298439 | 3.90 |
ENSDART00000170566
|
srpk3
|
SRSF protein kinase 3 |
| chr25_-_16818380 | 3.86 |
ENSDART00000155401
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr7_+_74134010 | 3.85 |
ENSDART00000164874
|
cldnd1a
|
claudin domain containing 1a |
| chr7_-_18881358 | 3.85 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr6_-_51101834 | 3.83 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr5_-_69923241 | 3.81 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr21_+_44300689 | 3.80 |
ENSDART00000186298
ENSDART00000142810 |
gabra3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr19_-_38872650 | 3.80 |
ENSDART00000146641
|
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr21_-_25601648 | 3.80 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr24_+_18714212 | 3.79 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr11_+_37049347 | 3.78 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr3_-_1204341 | 3.78 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr16_+_10963602 | 3.77 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr21_+_27382893 | 3.77 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr14_-_39074539 | 3.73 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr2_+_21309272 | 3.70 |
ENSDART00000141322
|
zbtb47a
|
zinc finger and BTB domain containing 47a |
| chr20_-_27325258 | 3.68 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr18_+_45550783 | 3.67 |
ENSDART00000138075
|
kifc3
|
kinesin family member C3 |
| chr11_-_11625630 | 3.63 |
ENSDART00000161821
ENSDART00000193152 |
si:dkey-28e7.3
|
si:dkey-28e7.3 |
| chr15_+_15516612 | 3.63 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr8_+_25254435 | 3.62 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr1_+_55294009 | 3.60 |
ENSDART00000128979
|
zgc:172106
|
zgc:172106 |
| chr3_+_38793499 | 3.59 |
ENSDART00000083394
|
kcnj19a
|
potassium voltage-gated channel subfamily J member 19a |
| chr15_-_31508221 | 3.58 |
ENSDART00000121464
|
si:dkey-1m11.5
|
si:dkey-1m11.5 |
| chr14_-_4556896 | 3.54 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr15_-_20954552 | 3.53 |
ENSDART00000006910
|
tbcela
|
tubulin folding cofactor E-like a |
| chr10_-_31440500 | 3.51 |
ENSDART00000024778
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr3_-_30885250 | 3.48 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
Network of associatons between targets according to the STRING database.
First level regulatory network of neurod1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.7 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 3.6 | 14.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 3.4 | 13.5 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 3.0 | 12.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 2.5 | 2.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 2.3 | 7.0 | GO:0050748 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 2.3 | 11.6 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 1.9 | 5.7 | GO:1901546 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 1.7 | 8.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 1.5 | 6.0 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 1.4 | 11.6 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 1.4 | 14.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.4 | 7.0 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 1.4 | 4.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.3 | 1.3 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 1.3 | 9.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 1.3 | 7.6 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 1.3 | 6.3 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.2 | 3.5 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 1.1 | 18.7 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 1.0 | 18.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.9 | 3.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.9 | 4.4 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.9 | 3.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.8 | 13.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.8 | 3.8 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.7 | 2.2 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.7 | 7.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.7 | 2.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.7 | 2.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.6 | 2.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.6 | 10.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.6 | 6.3 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.6 | 6.8 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.6 | 1.8 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.6 | 6.6 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.6 | 9.4 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.6 | 1.7 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.5 | 11.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.5 | 7.7 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.5 | 1.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.5 | 8.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.5 | 9.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 6.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.5 | 2.8 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 19.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.5 | 3.7 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.4 | 2.2 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.4 | 3.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.4 | 8.3 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.4 | 2.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.4 | 10.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.4 | 1.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.4 | 1.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.4 | 8.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.4 | 2.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.4 | 10.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.4 | 1.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.3 | 1.0 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.3 | 5.1 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.3 | 3.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 26.0 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 4.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.3 | 3.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.3 | 1.9 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.3 | 12.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.3 | 3.7 | GO:0001964 | startle response(GO:0001964) |
| 0.3 | 3.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.3 | 2.7 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.3 | 5.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.3 | 7.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.3 | 1.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 10.2 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.3 | 1.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 | 2.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.3 | 10.4 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.3 | 3.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.3 | 3.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) commissural neuron axon guidance(GO:0071679) |
| 0.3 | 11.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 0.7 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.2 | 4.1 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 1.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.2 | 1.6 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 2.6 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 1.2 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.2 | 6.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 1.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 2.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 9.3 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.2 | 6.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 2.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 3.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 0.6 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 0.8 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 0.2 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.2 | 1.9 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 0.5 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.2 | 3.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.8 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 3.1 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.2 | 7.2 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.2 | 5.3 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.2 | 3.8 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.2 | 2.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 6.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.2 | 9.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 1.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 0.9 | GO:0060997 | dendritic spine development(GO:0060996) dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 8.8 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 2.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.0 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) cellular response to leucine starvation(GO:1990253) |
| 0.1 | 5.7 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 3.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 21.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 16.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 1.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.6 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 1.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 7.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 2.1 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 2.9 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 3.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.5 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 1.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.8 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.1 | 1.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.1 | 3.2 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 1.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 2.9 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.8 | GO:0099536 | synaptic signaling(GO:0099536) |
| 0.1 | 3.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 3.7 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.1 | 0.7 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 3.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 0.4 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 26.9 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.1 | 2.0 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 20.9 | GO:0048812 | neuron projection morphogenesis(GO:0048812) |
| 0.1 | 1.0 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.8 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.1 | 1.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 0.7 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.0 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 1.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 3.3 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 19.9 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 6.8 | GO:0048511 | rhythmic process(GO:0048511) |
| 0.0 | 1.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.6 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 2.9 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 3.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 18.0 | GO:0030182 | neuron differentiation(GO:0030182) |
| 0.0 | 4.5 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 2.5 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 1.6 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.3 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 1.4 | GO:0060560 | developmental growth involved in morphogenesis(GO:0060560) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.9 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 2.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 3.3 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 1.4 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.4 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 1.1 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 4.2 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.1 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 5.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.9 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 17.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.4 | 11.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 1.4 | 5.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.4 | 5.6 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.9 | 10.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.8 | 9.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.8 | 6.0 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.7 | 7.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.7 | 12.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.7 | 4.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.6 | 1.9 | GO:1990745 | EARP complex(GO:1990745) |
| 0.6 | 3.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 9.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.5 | 1.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.5 | 16.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.5 | 8.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.4 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 41.0 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.4 | 6.8 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.3 | 12.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.3 | 9.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 9.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 1.9 | GO:0030891 | VCB complex(GO:0030891) |
| 0.3 | 2.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 2.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 6.1 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 1.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.3 | 6.2 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 2.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 1.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 0.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 1.2 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 11.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 3.8 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.2 | 2.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 2.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 1.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 10.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 15.1 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 9.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 5.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 4.3 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 12.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 13.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 0.4 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 5.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 37.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 4.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 3.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 10.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 4.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 9.7 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.1 | 0.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.7 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.1 | 18.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 2.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 4.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 8.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 3.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 3.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 2.5 | 22.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 2.2 | 15.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 2.2 | 11.1 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.8 | 9.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 1.7 | 41.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 1.6 | 18.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.5 | 7.5 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 1.4 | 9.9 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 1.2 | 3.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.1 | 10.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.0 | 3.1 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 1.0 | 13.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 1.0 | 3.0 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.9 | 4.4 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.9 | 2.6 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.8 | 6.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.8 | 14.0 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.8 | 6.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.8 | 6.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.8 | 36.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.8 | 5.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.7 | 2.1 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.7 | 2.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.7 | 6.6 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 1.8 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.6 | 7.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.6 | 8.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.6 | 2.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.6 | 8.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 2.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 3.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.5 | 8.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.5 | 4.3 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.5 | 8.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.5 | 2.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.4 | 1.7 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.4 | 12.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 3.7 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.4 | 17.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.4 | 2.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 2.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.4 | 2.9 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 4.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 3.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 10.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 8.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 4.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 3.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 9.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 4.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 3.8 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.3 | 6.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 9.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 2.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.2 | 6.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 1.7 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 1.2 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.2 | 5.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 2.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 5.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.7 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.2 | 0.9 | GO:0042903 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 3.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 8.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 1.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 2.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 4.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 5.8 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.2 | 0.8 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 17.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 4.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.2 | 9.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 1.6 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 1.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 1.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 11.1 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 2.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 3.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 2.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 65.2 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 2.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 6.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 4.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.4 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.0 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 4.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 6.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 1.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.9 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.1 | 1.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 4.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.1 | 2.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 2.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 3.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 2.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 3.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 5.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 17.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 4.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 2.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 3.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 2.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 28.4 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 5.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 3.3 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.0 | 2.7 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 7.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 11.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 14.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 15.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 5.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 1.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 2.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 6.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.2 | 5.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 4.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 2.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.1 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 2.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 1.7 | 18.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.9 | 16.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.8 | 10.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 7.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 3.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 4.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.4 | 3.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 7.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 5.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 3.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 4.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 1.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.3 | 7.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.3 | 4.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 4.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 17.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 16.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.6 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.2 | 1.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 4.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 3.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 1.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.1 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 2.4 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |