Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
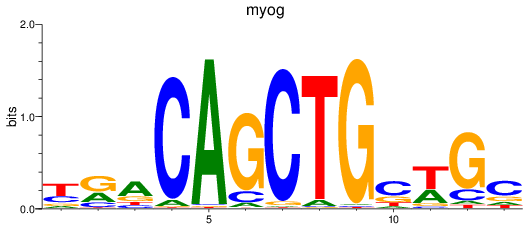
Results for myog
Z-value: 1.31
Transcription factors associated with myog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myog
|
ENSDARG00000009438 | myogenin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myog | dr11_v1_chr11_-_22599584_22599584 | 0.59 | 5.3e-10 | Click! |
Activity profile of myog motif
Sorted Z-values of myog motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61203203 | 29.67 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr21_+_27382893 | 24.80 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr7_+_29951997 | 22.39 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr20_-_27325258 | 18.30 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr18_+_5549672 | 14.10 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr2_+_42191592 | 14.06 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr23_-_45405968 | 13.89 |
ENSDART00000149462
|
zgc:101853
|
zgc:101853 |
| chr7_+_39444843 | 13.67 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr3_+_26145013 | 11.88 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr24_+_20575259 | 11.39 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr22_+_20720808 | 11.29 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr6_-_32703317 | 11.22 |
ENSDART00000064833
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr8_-_1051438 | 10.34 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr2_-_21335131 | 10.33 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr11_-_5865744 | 9.63 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr6_-_21492752 | 9.36 |
ENSDART00000006843
ENSDART00000171479 |
cacng1a
|
calcium channel, voltage-dependent, gamma subunit 1a |
| chr16_-_14074594 | 9.30 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr6_-_42003780 | 9.29 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr9_-_100579 | 9.13 |
ENSDART00000006099
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr19_-_10425140 | 8.95 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr11_+_7158723 | 8.92 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr13_-_31622195 | 8.65 |
ENSDART00000057432
|
six1a
|
SIX homeobox 1a |
| chr18_+_5454341 | 8.49 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr24_-_33703504 | 7.99 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr3_+_57991074 | 7.90 |
ENSDART00000076077
|
myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr19_+_14109348 | 7.39 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr3_+_26144765 | 7.34 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr17_+_27434626 | 7.30 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr9_-_48281941 | 6.82 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr16_-_42894628 | 6.81 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr14_-_9281232 | 6.70 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr20_-_36887516 | 6.64 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr10_-_44560165 | 6.59 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr23_+_44614056 | 6.56 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr17_-_722218 | 6.46 |
ENSDART00000160385
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr18_+_23218980 | 6.42 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr23_-_18057553 | 6.28 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr9_-_43071519 | 6.27 |
ENSDART00000109099
|
ttn.2
|
titin, tandem duplicate 2 |
| chr24_+_39108243 | 6.20 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr17_-_14815557 | 6.15 |
ENSDART00000154473
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr6_-_32093830 | 5.83 |
ENSDART00000017695
|
foxd3
|
forkhead box D3 |
| chr23_+_22656477 | 5.73 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr20_-_54381034 | 5.58 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr21_-_30254185 | 5.48 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr20_-_36887176 | 5.46 |
ENSDART00000143515
ENSDART00000146994 |
txlnba
|
taxilin beta a |
| chr16_+_54588930 | 5.45 |
ENSDART00000159174
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr20_+_26967154 | 5.34 |
ENSDART00000153294
ENSDART00000132434 ENSDART00000143047 |
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr23_-_18057270 | 5.30 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr8_+_44926946 | 5.29 |
ENSDART00000098567
|
zgc:154046
|
zgc:154046 |
| chr4_-_2350371 | 5.28 |
ENSDART00000166274
|
phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr9_-_30145080 | 4.99 |
ENSDART00000133746
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr23_-_18057851 | 4.92 |
ENSDART00000173075
ENSDART00000173230 ENSDART00000173135 ENSDART00000173431 ENSDART00000173068 ENSDART00000172987 |
zgc:92287
|
zgc:92287 |
| chr8_-_11229523 | 4.85 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr23_-_32162810 | 4.77 |
ENSDART00000155905
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr19_+_43119698 | 4.71 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr11_-_45171139 | 4.68 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr2_+_50999477 | 4.46 |
ENSDART00000190111
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr6_+_40629066 | 4.45 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr12_+_6002715 | 4.39 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr22_-_10470663 | 4.38 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr7_-_20582842 | 4.37 |
ENSDART00000169750
ENSDART00000111719 |
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr19_+_30990815 | 4.35 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr8_-_51404806 | 4.32 |
ENSDART00000060625
|
lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr16_-_31469065 | 4.30 |
ENSDART00000182397
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr17_-_22001303 | 4.29 |
ENSDART00000122190
|
slc22a7b.2
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 2 |
| chr2_+_6253246 | 4.16 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr1_-_58868306 | 4.07 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr21_-_5056812 | 4.06 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr1_-_471925 | 4.00 |
ENSDART00000152684
|
zgc:92518
|
zgc:92518 |
| chr20_+_23625387 | 3.98 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr9_-_68934 | 3.97 |
ENSDART00000054594
ENSDART00000009389 |
il10rb
|
interleukin 10 receptor, beta |
| chr17_+_132555 | 3.94 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr23_+_553396 | 3.90 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr17_-_34963575 | 3.90 |
ENSDART00000145664
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr23_-_31633201 | 3.78 |
ENSDART00000143335
ENSDART00000053531 |
slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr18_+_23249519 | 3.77 |
ENSDART00000005740
ENSDART00000147446 ENSDART00000124818 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr25_+_17689565 | 3.59 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr9_-_7655243 | 3.57 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr4_+_10066840 | 3.53 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr7_+_31838320 | 3.52 |
ENSDART00000144679
ENSDART00000174217 ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr25_-_29611476 | 3.49 |
ENSDART00000154458
|
si:ch211-253p14.2
|
si:ch211-253p14.2 |
| chr3_-_62380146 | 3.48 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr2_+_32016516 | 3.44 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr13_+_228045 | 3.38 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr25_+_13406069 | 3.36 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr14_+_7939398 | 3.29 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr17_-_14836320 | 3.27 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr21_-_217589 | 3.11 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr6_-_10964083 | 3.10 |
ENSDART00000181583
|
notum2
|
notum pectinacetylesterase 2 |
| chr18_+_15271993 | 3.09 |
ENSDART00000099777
|
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr25_+_16214854 | 3.08 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr8_+_4333914 | 3.04 |
ENSDART00000137528
|
myl2b
|
myosin, light chain 2b, regulatory, cardiac, slow |
| chr14_-_32258759 | 3.01 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr14_-_29906209 | 2.93 |
ENSDART00000192952
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr5_-_48307804 | 2.85 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr14_+_7939216 | 2.82 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr3_-_50865079 | 2.81 |
ENSDART00000164295
|
pmp22a
|
peripheral myelin protein 22a |
| chr2_-_17115256 | 2.81 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr1_-_58880613 | 2.76 |
ENSDART00000188136
|
CABZ01084501.2
|
|
| chr17_-_27048537 | 2.76 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr1_+_27690 | 2.71 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr18_-_14274803 | 2.68 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr16_+_14010242 | 2.67 |
ENSDART00000059928
|
fdps
|
farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) |
| chr2_+_50999675 | 2.66 |
ENSDART00000158064
ENSDART00000165746 ENSDART00000163917 ENSDART00000172038 ENSDART00000169048 ENSDART00000164775 |
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr4_-_18211 | 2.64 |
ENSDART00000171737
|
ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr11_-_101758 | 2.61 |
ENSDART00000173015
|
elmo2
|
engulfment and cell motility 2 |
| chr25_-_17587785 | 2.59 |
ENSDART00000073679
ENSDART00000146851 |
zgc:66449
|
zgc:66449 |
| chr7_-_73856073 | 2.56 |
ENSDART00000181793
|
FP236812.2
|
|
| chr6_+_153146 | 2.52 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr5_+_4806851 | 2.51 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr16_+_54592907 | 2.50 |
ENSDART00000172113
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr10_+_24468922 | 2.49 |
ENSDART00000008248
ENSDART00000183510 |
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr11_-_55224 | 2.48 |
ENSDART00000159169
|
col2a1b
|
collagen, type II, alpha 1b |
| chr7_+_67381912 | 2.46 |
ENSDART00000167564
|
nfat5b
|
nuclear factor of activated T cells 5b |
| chr3_-_1388936 | 2.46 |
ENSDART00000171278
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr8_+_54284961 | 2.45 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr22_-_968484 | 2.45 |
ENSDART00000105895
|
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr9_+_22657221 | 2.40 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr1_+_8508753 | 2.37 |
ENSDART00000091683
|
alkbh5
|
alkB homolog 5, RNA demethylase |
| chr4_-_77624155 | 2.34 |
ENSDART00000099761
|
si:ch211-250m6.7
|
si:ch211-250m6.7 |
| chr23_+_43668756 | 2.31 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
| chr5_+_51226846 | 2.29 |
ENSDART00000138210
|
ubac1
|
UBA domain containing 1 |
| chr14_-_30050 | 2.28 |
ENSDART00000164411
|
zbtb49
|
zinc finger and BTB domain containing 49 |
| chr15_-_12500938 | 2.27 |
ENSDART00000159627
|
scn4ba
|
sodium channel, voltage-gated, type IV, beta a |
| chr7_+_46003449 | 2.23 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr8_-_46386024 | 2.22 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr2_-_17114852 | 2.20 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr2_-_2937225 | 2.20 |
ENSDART00000168132
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr7_-_58130703 | 2.18 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr12_-_4756478 | 2.14 |
ENSDART00000152181
|
mapta
|
microtubule-associated protein tau a |
| chr10_+_7636811 | 2.14 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr12_-_11570 | 2.14 |
ENSDART00000186179
|
shisa6
|
shisa family member 6 |
| chr22_-_8509215 | 2.13 |
ENSDART00000140146
|
si:ch73-27e22.3
|
si:ch73-27e22.3 |
| chr13_-_214122 | 2.13 |
ENSDART00000169273
|
ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr2_+_258698 | 2.12 |
ENSDART00000181330
ENSDART00000181645 |
phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr2_-_42393590 | 2.11 |
ENSDART00000135529
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr20_+_34537736 | 2.10 |
ENSDART00000152982
|
si:ch211-242b18.1
|
si:ch211-242b18.1 |
| chr25_+_35683956 | 2.09 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr7_-_74090168 | 2.07 |
ENSDART00000050528
|
tyrp1a
|
tyrosinase-related protein 1a |
| chr19_-_2420990 | 2.04 |
ENSDART00000181498
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr3_+_7771420 | 2.04 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr2_+_32016256 | 2.03 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr9_+_22656976 | 2.03 |
ENSDART00000136249
ENSDART00000139270 |
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr23_+_45845159 | 2.00 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr5_+_1515938 | 1.97 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr8_+_26410539 | 1.94 |
ENSDART00000168780
|
ifrd2
|
interferon-related developmental regulator 2 |
| chr17_+_53296851 | 1.94 |
ENSDART00000158313
|
ddx24
|
DEAD (Asp-Glu-Ala-Asp) box helicase 24 |
| chr2_+_29995590 | 1.92 |
ENSDART00000151906
|
rbm33b
|
RNA binding motif protein 33b |
| chr8_-_8446668 | 1.92 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr15_-_39969988 | 1.89 |
ENSDART00000146054
|
rps5
|
ribosomal protein S5 |
| chr25_-_20258508 | 1.89 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr19_+_15441022 | 1.88 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr22_-_29336268 | 1.87 |
ENSDART00000132776
ENSDART00000186351 ENSDART00000121599 |
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr12_+_15165736 | 1.86 |
ENSDART00000180398
|
adprh
|
ADP-ribosylarginine hydrolase |
| chr19_-_46957968 | 1.86 |
ENSDART00000043713
|
angpt1
|
angiopoietin 1 |
| chr12_-_26415499 | 1.85 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr7_+_16963091 | 1.85 |
ENSDART00000173770
|
nav2a
|
neuron navigator 2a |
| chr13_-_32726178 | 1.84 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr7_+_5911670 | 1.82 |
ENSDART00000173025
|
si:dkey-23a13.9
|
si:dkey-23a13.9 |
| chr2_+_47708853 | 1.81 |
ENSDART00000124307
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr5_-_32336613 | 1.81 |
ENSDART00000139732
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr23_-_27505825 | 1.80 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr25_-_3549321 | 1.78 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr9_-_41025062 | 1.78 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr10_+_45059702 | 1.77 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| chr4_+_3980247 | 1.74 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr3_+_57788761 | 1.74 |
ENSDART00000149741
|
si:ch73-362i18.1
|
si:ch73-362i18.1 |
| chr8_-_43677762 | 1.71 |
ENSDART00000167762
|
ep400
|
E1A binding protein p400 |
| chr5_+_7564644 | 1.71 |
ENSDART00000192173
|
CABZ01039096.1
|
|
| chr12_-_10508952 | 1.68 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr4_-_5597802 | 1.67 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr14_+_36497250 | 1.66 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr18_-_17724295 | 1.66 |
ENSDART00000121553
|
fam192a
|
family with sequence similarity 192, member A |
| chr4_-_8093753 | 1.64 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr8_+_1284784 | 1.63 |
ENSDART00000061663
|
fbxl17
|
F-box and leucine-rich repeat protein 17 |
| chr8_-_45838481 | 1.63 |
ENSDART00000148206
ENSDART00000170485 |
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr11_+_7214353 | 1.62 |
ENSDART00000156764
|
nwd1
|
NACHT and WD repeat domain containing 1 |
| chr6_-_9952103 | 1.62 |
ENSDART00000065475
|
zp2l2
|
zona pellucida glycoprotein 2, like 2 |
| chr2_-_7666021 | 1.61 |
ENSDART00000180007
|
CABZ01021592.1
|
|
| chr18_-_10995410 | 1.61 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr23_-_1583193 | 1.61 |
ENSDART00000143841
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr13_-_24745288 | 1.60 |
ENSDART00000031564
|
sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
| chr3_+_17251542 | 1.60 |
ENSDART00000024832
|
stat5a
|
signal transducer and activator of transcription 5a |
| chr5_+_6796291 | 1.57 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr9_+_51891 | 1.55 |
ENSDART00000163529
|
zgc:158316
|
zgc:158316 |
| chr10_-_690072 | 1.54 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr8_-_45838277 | 1.54 |
ENSDART00000046064
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr25_-_36365977 | 1.52 |
ENSDART00000152546
|
zgc:173552
|
zgc:173552 |
| chr12_-_49166761 | 1.52 |
ENSDART00000189407
|
acadsb
|
acyl-CoA dehydrogenase short/branched chain |
| chr8_-_26609259 | 1.50 |
ENSDART00000027301
|
sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr14_+_21828993 | 1.50 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr7_-_6368406 | 1.50 |
ENSDART00000172787
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr8_+_26410197 | 1.50 |
ENSDART00000145836
ENSDART00000053447 |
ifrd2
|
interferon-related developmental regulator 2 |
| chr16_-_41059919 | 1.50 |
ENSDART00000188803
|
si:dkey-201i6.8
|
si:dkey-201i6.8 |
| chr16_+_41873708 | 1.49 |
ENSDART00000084631
ENSDART00000084639 ENSDART00000058611 |
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr7_-_41468751 | 1.48 |
ENSDART00000150146
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr20_+_46268906 | 1.48 |
ENSDART00000113372
|
taar14e
|
trace amine associated receptor 14e |
| chr9_-_17783574 | 1.47 |
ENSDART00000146706
|
vwa8
|
von Willebrand factor A domain containing 8 |
| chr7_-_69636502 | 1.47 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr18_+_40584288 | 1.47 |
ENSDART00000087692
|
si:ch211-132b12.3
|
si:ch211-132b12.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of myog
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.7 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 4.8 | 19.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 3.5 | 14.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 3.2 | 9.6 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 2.2 | 8.6 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 1.9 | 9.4 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 1.5 | 12.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 1.4 | 12.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 1.4 | 24.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 1.2 | 5.8 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 1.1 | 8.0 | GO:0033292 | T-tubule organization(GO:0033292) |
| 1.1 | 9.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 1.1 | 5.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.0 | 16.6 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.8 | 3.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.8 | 15.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.7 | 1.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.7 | 3.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.6 | 2.4 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.6 | 2.3 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.5 | 2.7 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.5 | 8.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.5 | 3.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.5 | 4.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.5 | 5.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.5 | 3.8 | GO:0046323 | glucose import(GO:0046323) |
| 0.5 | 1.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.5 | 7.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.4 | 2.7 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.4 | 6.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.4 | 1.3 | GO:0019264 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.4 | 5.0 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.4 | 2.0 | GO:1990564 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 1.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 1.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.4 | 1.1 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.4 | 1.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.3 | 2.4 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.3 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 5.6 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.3 | 1.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.3 | 4.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.3 | 1.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.3 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.3 | 2.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 1.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.3 | 1.3 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.3 | 2.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 0.7 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 4.2 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 1.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 2.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 2.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.7 | GO:0044406 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.2 | 1.8 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.2 | 9.4 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.2 | 22.4 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.2 | 1.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 0.6 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.2 | 1.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 8.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.2 | 1.2 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.2 | 1.5 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.2 | 8.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.2 | 0.7 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.2 | 1.5 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.2 | 2.3 | GO:2000651 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 2.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 4.4 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.2 | 0.9 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 1.7 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.2 | 2.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.7 | GO:0016037 | light absorption(GO:0016037) |
| 0.1 | 1.3 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 9.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 1.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.9 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:1901380 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.8 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 1.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 4.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 2.8 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.1 | 11.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 6.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 0.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 2.4 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.1 | 4.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.1 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.1 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.6 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 8.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.8 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 2.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 1.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.5 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.8 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.8 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 1.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 1.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 9.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 5.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.9 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 1.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 2.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.0 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 2.8 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 0.3 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.1 | 0.5 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 3.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.9 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.5 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.1 | 0.2 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.0 | 0.3 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 3.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.8 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 1.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 1.3 | GO:0009261 | purine ribonucleotide catabolic process(GO:0009154) ribonucleotide catabolic process(GO:0009261) |
| 0.0 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.7 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 1.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 24.2 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.8 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 2.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 1.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.5 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.6 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 2.3 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.6 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 2.6 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 1.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 3.4 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 3.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.5 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 2.0 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 3.2 | GO:0043410 | positive regulation of MAPK cascade(GO:0043410) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.3 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 0.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 2.6 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.8 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 19.2 | GO:0031673 | H zone(GO:0031673) |
| 1.5 | 12.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.4 | 36.9 | GO:0031672 | A band(GO:0031672) |
| 1.3 | 9.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.0 | 7.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.7 | 15.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.7 | 31.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.6 | 3.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.4 | 2.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 13.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 3.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 1.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 2.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 0.7 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.2 | 3.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.6 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.2 | 5.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 22.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 1.8 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.2 | 2.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 1.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 15.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 2.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 2.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.5 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 2.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 2.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 9.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 12.1 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.1 | 4.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 1.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 1.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 21.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 7.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.2 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 2.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 3.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 3.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.1 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 5.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 6.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 5.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 14.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 2.3 | 9.3 | GO:0071253 | connexin binding(GO:0071253) |
| 1.5 | 12.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.4 | 5.7 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 1.3 | 4.0 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 1.2 | 13.7 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 1.1 | 19.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 1.0 | 5.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 1.0 | 14.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.9 | 2.7 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.8 | 3.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.6 | 2.4 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.5 | 2.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.5 | 3.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.5 | 6.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.5 | 5.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.5 | 5.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 8.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 1.3 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.4 | 1.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.4 | 10.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.4 | 4.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 3.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 1.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 4.2 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.4 | 1.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.3 | 7.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.3 | 3.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 2.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 4.9 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 2.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.3 | 13.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.3 | 0.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 1.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 6.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 10.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 1.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 3.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 0.8 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.2 | 2.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 0.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.7 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.2 | 0.9 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.2 | 0.9 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.2 | 22.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.2 | 0.5 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 1.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 2.3 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.7 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 10.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 3.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 2.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 12.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 4.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 1.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 3.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.8 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 2.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.7 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 12.2 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 3.3 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 9.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 4.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 3.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 17.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 1.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 3.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 9.1 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 4.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.5 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 7.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 3.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 1.8 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 1.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 7.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 1.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 2.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 11.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0019955 | cytokine binding(GO:0019955) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 4.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 9.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 7.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.2 | 6.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 5.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.5 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 19.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.6 | 6.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 3.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 6.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 1.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 1.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 6.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 2.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 3.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 8.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.9 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 3.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |