Project
PRJDB7713: Age associated transcriptome analysis in 5 tissues of zebrafish
Navigation
Downloads
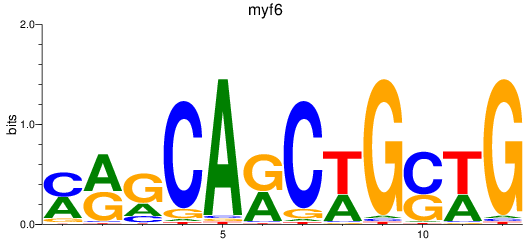
Results for myf6
Z-value: 1.47
Transcription factors associated with myf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myf6
|
ENSDARG00000029830 | myogenic factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myf6 | dr11_v1_chr4_+_21717793_21717793 | 0.46 | 3.1e-06 | Click! |
Activity profile of myf6 motif
Sorted Z-values of myf6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_13256415 | 10.70 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr9_-_105135 | 10.17 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr2_+_42191592 | 10.17 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr19_-_5332784 | 6.86 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr18_+_1615 | 6.80 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr13_-_214122 | 5.37 |
ENSDART00000169273
|
ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr13_-_280652 | 5.14 |
ENSDART00000193627
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr18_-_50956280 | 4.83 |
ENSDART00000058457
|
st7
|
suppression of tumorigenicity 7 |
| chr3_-_61203203 | 4.72 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr22_+_20720808 | 4.68 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr9_+_51891 | 4.63 |
ENSDART00000163529
|
zgc:158316
|
zgc:158316 |
| chr9_-_68934 | 4.49 |
ENSDART00000054594
ENSDART00000009389 |
il10rb
|
interleukin 10 receptor, beta |
| chr3_+_30257582 | 4.47 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr4_+_90048 | 4.30 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr25_+_245018 | 4.21 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr21_-_45891262 | 4.20 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr3_+_26145013 | 4.19 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr7_+_1467863 | 4.09 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr13_+_233482 | 4.06 |
ENSDART00000102511
|
cfap36
|
cilia and flagella associated protein 36 |
| chr14_+_97017 | 3.99 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr12_+_18782821 | 3.92 |
ENSDART00000152918
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr3_-_1388936 | 3.89 |
ENSDART00000171278
|
ddx47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr23_+_2714949 | 3.86 |
ENSDART00000105284
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr1_+_218524 | 3.86 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr10_+_6318227 | 3.81 |
ENSDART00000170872
ENSDART00000162428 ENSDART00000158994 |
tpm2
|
tropomyosin 2 (beta) |
| chr21_+_27382893 | 3.74 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr5_-_71705191 | 3.65 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr13_-_280827 | 3.55 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr1_+_18716 | 3.54 |
ENSDART00000172454
ENSDART00000161190 |
nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr2_-_44280061 | 3.54 |
ENSDART00000136818
|
mpz
|
myelin protein zero |
| chr22_-_37738203 | 3.48 |
ENSDART00000143190
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr24_-_33703504 | 3.45 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr2_+_68789 | 3.44 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr8_+_54202554 | 3.43 |
ENSDART00000020569
|
creld1b
|
cysteine-rich with EGF-like domains 1b |
| chr17_+_132555 | 3.42 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr21_-_131236 | 3.38 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr1_+_27690 | 3.37 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr21_-_217589 | 3.32 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr3_+_26144765 | 3.19 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr1_-_45177373 | 3.18 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr1_-_58059134 | 3.16 |
ENSDART00000160970
|
caspb
|
caspase b |
| chr24_-_27473771 | 3.11 |
ENSDART00000139874
|
cxl34b.11
|
CX chemokine ligand 34b, duplicate 11 |
| chr14_+_52369262 | 3.09 |
ENSDART00000169352
ENSDART00000157833 |
igfbp7
|
insulin-like growth factor binding protein 7 |
| chr9_-_216527 | 2.97 |
ENSDART00000163068
|
aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr15_-_2652640 | 2.96 |
ENSDART00000146094
|
cldnf
|
claudin f |
| chr23_+_6077503 | 2.96 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr11_-_45171139 | 2.92 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr25_+_37435720 | 2.87 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr1_-_69444 | 2.85 |
ENSDART00000166954
|
si:zfos-1011f11.1
|
si:zfos-1011f11.1 |
| chr1_+_52398205 | 2.84 |
ENSDART00000143225
|
si:ch211-217k17.9
|
si:ch211-217k17.9 |
| chr12_-_212843 | 2.82 |
ENSDART00000083574
|
CABZ01102039.1
|
|
| chr20_-_27325258 | 2.81 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr19_+_551963 | 2.81 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr23_+_1349277 | 2.77 |
ENSDART00000173133
ENSDART00000179877 |
utrn
|
utrophin |
| chr25_+_4581214 | 2.74 |
ENSDART00000185552
|
CABZ01068600.1
|
|
| chr3_-_28075756 | 2.73 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr5_-_24201437 | 2.73 |
ENSDART00000114113
|
sox19a
|
SRY (sex determining region Y)-box 19a |
| chr5_-_417495 | 2.72 |
ENSDART00000180586
ENSDART00000189408 |
HOOK3
|
hook microtubule tethering protein 3 |
| chr19_-_30810328 | 2.70 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr1_+_55002583 | 2.70 |
ENSDART00000037250
|
si:ch211-196h16.12
|
si:ch211-196h16.12 |
| chr25_-_37489917 | 2.69 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr7_+_25323742 | 2.67 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr4_-_64703 | 2.66 |
ENSDART00000167851
|
CU856344.1
|
|
| chr21_+_13387965 | 2.66 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr17_-_7861219 | 2.65 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr2_-_44255537 | 2.61 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr9_-_100579 | 2.60 |
ENSDART00000006099
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr15_-_47458034 | 2.60 |
ENSDART00000168527
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr8_+_53344726 | 2.59 |
ENSDART00000184395
ENSDART00000170212 |
CU914536.1
|
|
| chr13_-_438705 | 2.59 |
ENSDART00000082142
|
CU570800.1
|
|
| chr14_-_41388178 | 2.53 |
ENSDART00000124532
ENSDART00000125016 ENSDART00000169247 |
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr12_-_961014 | 2.51 |
ENSDART00000088351
ENSDART00000180866 ENSDART00000186695 ENSDART00000190850 |
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr20_-_2361226 | 2.51 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr22_+_19188809 | 2.48 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr2_-_57837838 | 2.47 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr13_+_9100 | 2.46 |
ENSDART00000165772
|
ppp4r3b
|
protein phosphatase 4, regulatory subunit 3B |
| chr24_+_42132962 | 2.46 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr8_-_50259448 | 2.45 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr16_+_20738740 | 2.42 |
ENSDART00000079343
|
jazf1b
|
JAZF zinc finger 1b |
| chr7_-_22632690 | 2.42 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr11_+_45448212 | 2.41 |
ENSDART00000173341
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr11_+_31558207 | 2.38 |
ENSDART00000140204
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr20_-_54377933 | 2.38 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr25_+_186583 | 2.36 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr2_+_58739111 | 2.34 |
ENSDART00000097690
|
CABZ01083448.1
|
|
| chr8_-_26792912 | 2.31 |
ENSDART00000139787
|
kazna
|
kazrin, periplakin interacting protein a |
| chr21_+_30950097 | 2.28 |
ENSDART00000187572
|
rhogb
|
ras homolog family member Gb |
| chr17_-_51224159 | 2.23 |
ENSDART00000185749
|
psen1
|
presenilin 1 |
| chr12_+_27331324 | 2.22 |
ENSDART00000087208
|
sost
|
sclerostin |
| chr5_-_64203101 | 2.20 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr24_-_42090635 | 2.20 |
ENSDART00000166413
|
ssr1
|
signal sequence receptor, alpha |
| chr15_+_509126 | 2.17 |
ENSDART00000102274
|
ftr86
|
finTRIM family, member 86 |
| chr15_+_47746176 | 2.16 |
ENSDART00000154481
ENSDART00000160914 |
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr6_+_42820228 | 2.15 |
ENSDART00000185413
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr19_-_48010490 | 2.15 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr4_+_38344 | 2.13 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr22_+_987788 | 2.12 |
ENSDART00000149486
|
def6b
|
differentially expressed in FDCP 6b homolog (mouse) |
| chr19_+_48060464 | 2.11 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr15_+_47161917 | 2.10 |
ENSDART00000167860
|
gap43
|
growth associated protein 43 |
| chr13_-_2981472 | 2.09 |
ENSDART00000184820
|
CABZ01087623.1
|
|
| chr20_-_40717900 | 2.09 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr18_+_15876385 | 2.06 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr19_+_43037657 | 2.05 |
ENSDART00000168263
ENSDART00000184771 ENSDART00000164453 ENSDART00000165202 |
pum1
|
pumilio RNA-binding family member 1 |
| chr1_+_54124209 | 2.04 |
ENSDART00000187730
|
LO017722.1
|
|
| chr7_+_49715750 | 2.03 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr11_-_45370296 | 2.02 |
ENSDART00000168770
|
trappc10
|
trafficking protein particle complex 10 |
| chr7_+_30988570 | 1.99 |
ENSDART00000180613
ENSDART00000185625 |
tjp1a
|
tight junction protein 1a |
| chr10_-_20357013 | 1.99 |
ENSDART00000080143
|
sfrp1b
|
secreted frizzled-related protein 1b |
| chr19_-_10771558 | 1.98 |
ENSDART00000085165
|
slc9a3.2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3, tandem duplicate 2 |
| chr24_-_42108655 | 1.97 |
ENSDART00000032722
|
pigm
|
phosphatidylinositol glycan anchor biosynthesis, class M |
| chr20_+_22681066 | 1.97 |
ENSDART00000143286
|
lnx1
|
ligand of numb-protein X 1 |
| chr12_-_46985 | 1.96 |
ENSDART00000152327
|
wdr45b
|
WD repeat domain 45B |
| chr25_+_22853718 | 1.94 |
ENSDART00000073588
|
kcnj11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr16_-_14074594 | 1.93 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr10_-_8197049 | 1.92 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr23_+_43177290 | 1.92 |
ENSDART00000193300
ENSDART00000186065 |
si:dkey-65j6.2
|
si:dkey-65j6.2 |
| chr17_-_11329959 | 1.91 |
ENSDART00000015418
|
irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr15_+_59417 | 1.91 |
ENSDART00000187260
|
AXL
|
AXL receptor tyrosine kinase |
| chr1_-_156375 | 1.90 |
ENSDART00000160221
|
pcid2
|
PCI domain containing 2 |
| chr6_+_42819337 | 1.88 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr12_-_44148073 | 1.88 |
ENSDART00000166273
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr21_+_19925910 | 1.87 |
ENSDART00000111694
ENSDART00000132653 |
tnksa
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase a |
| chr24_+_42149453 | 1.86 |
ENSDART00000128766
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr13_-_4848889 | 1.86 |
ENSDART00000165259
|
mcu
|
mitochondrial calcium uniporter |
| chr3_+_1211242 | 1.83 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr11_-_30636163 | 1.82 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr9_-_98982 | 1.82 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr18_+_22504200 | 1.81 |
ENSDART00000184472
ENSDART00000176130 ENSDART00000183780 ENSDART00000190379 ENSDART00000192281 |
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr7_+_3357973 | 1.81 |
ENSDART00000172853
|
si:ch211-285c6.5
|
si:ch211-285c6.5 |
| chr7_-_62003831 | 1.79 |
ENSDART00000113585
ENSDART00000062704 |
plaa
|
phospholipase A2-activating protein |
| chr10_-_309894 | 1.78 |
ENSDART00000163287
|
CABZ01049607.1
|
|
| chr11_-_101758 | 1.78 |
ENSDART00000173015
|
elmo2
|
engulfment and cell motility 2 |
| chr8_-_36140405 | 1.77 |
ENSDART00000182806
|
CT583723.2
|
|
| chr10_-_44306399 | 1.77 |
ENSDART00000180042
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
| chr2_-_57891504 | 1.77 |
ENSDART00000139948
|
zgc:92789
|
zgc:92789 |
| chr20_-_2584101 | 1.76 |
ENSDART00000141595
ENSDART00000135760 |
med23
|
mediator complex subunit 23 |
| chr14_-_246342 | 1.76 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr5_-_24712405 | 1.75 |
ENSDART00000033630
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr14_+_146857 | 1.75 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr1_-_25496133 | 1.75 |
ENSDART00000102498
|
sfrp2
|
secreted frizzled-related protein 2 |
| chr17_+_15535501 | 1.73 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr8_-_8446668 | 1.72 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr5_+_43530388 | 1.72 |
ENSDART00000190254
ENSDART00000097618 ENSDART00000133006 |
si:dkey-40c11.2
|
si:dkey-40c11.2 |
| chr21_+_6613772 | 1.72 |
ENSDART00000159645
|
col5a1
|
procollagen, type V, alpha 1 |
| chr3_+_59851537 | 1.71 |
ENSDART00000180997
|
CU693479.1
|
|
| chr12_-_44016898 | 1.71 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr1_+_58990121 | 1.70 |
ENSDART00000171654
|
CABZ01083166.1
|
|
| chr11_-_12233 | 1.70 |
ENSDART00000173352
ENSDART00000173009 ENSDART00000102293 |
myg1
|
melanocyte proliferating gene 1 |
| chr9_-_29029643 | 1.70 |
ENSDART00000151941
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr20_+_52774730 | 1.69 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr24_+_41989108 | 1.69 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr19_-_35400819 | 1.69 |
ENSDART00000148080
|
rnf19b
|
ring finger protein 19B |
| chr6_-_6976096 | 1.69 |
ENSDART00000151822
ENSDART00000039443 ENSDART00000177960 |
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr16_-_19568795 | 1.68 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr21_-_3452683 | 1.68 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr8_-_425255 | 1.67 |
ENSDART00000168496
ENSDART00000162614 |
fem1c
|
fem-1 homolog c (C. elegans) |
| chr7_-_20756013 | 1.67 |
ENSDART00000185259
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr21_+_25071805 | 1.66 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr23_+_37579107 | 1.66 |
ENSDART00000169376
|
plekhg5b
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5b |
| chr11_-_44999858 | 1.63 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr25_+_17871089 | 1.63 |
ENSDART00000133725
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr8_+_26522013 | 1.63 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr12_-_44010532 | 1.63 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr17_-_5769196 | 1.62 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr9_+_32872690 | 1.62 |
ENSDART00000020798
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr9_-_41784799 | 1.62 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr3_-_2613990 | 1.62 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr12_+_466324 | 1.62 |
ENSDART00000169691
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr11_+_25276748 | 1.61 |
ENSDART00000126211
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr18_-_210478 | 1.61 |
ENSDART00000136693
|
tm2d3
|
TM2 domain containing 3 |
| chr5_+_26212621 | 1.61 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr9_+_56443416 | 1.60 |
ENSDART00000167947
|
CABZ01079480.1
|
|
| chr24_-_1657276 | 1.59 |
ENSDART00000168131
|
si:ch73-378g22.1
|
si:ch73-378g22.1 |
| chr24_-_33291784 | 1.58 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr20_-_54381034 | 1.58 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr16_+_33931032 | 1.57 |
ENSDART00000167240
|
snip1
|
Smad nuclear interacting protein |
| chr6_+_12006557 | 1.57 |
ENSDART00000128024
|
wdsub1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr18_+_54354 | 1.57 |
ENSDART00000097163
|
zgc:158482
|
zgc:158482 |
| chr1_-_48933 | 1.56 |
ENSDART00000171162
|
ildr1a
|
immunoglobulin-like domain containing receptor 1a |
| chr21_+_28958471 | 1.56 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr19_+_32979132 | 1.55 |
ENSDART00000169469
ENSDART00000171782 ENSDART00000180705 ENSDART00000179326 ENSDART00000193791 |
spire1a
|
spire-type actin nucleation factor 1a |
| chr7_+_41147732 | 1.55 |
ENSDART00000185388
|
puf60b
|
poly-U binding splicing factor b |
| chr23_-_12423778 | 1.55 |
ENSDART00000124091
|
wfdc2
|
WAP four-disulfide core domain 2 |
| chr24_-_5691956 | 1.55 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr13_-_3324764 | 1.54 |
ENSDART00000102748
ENSDART00000114040 |
ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr23_-_27505825 | 1.54 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr7_+_34506937 | 1.53 |
ENSDART00000111303
|
rfx7a
|
regulatory factor X7a |
| chr14_-_30050 | 1.53 |
ENSDART00000164411
|
zbtb49
|
zinc finger and BTB domain containing 49 |
| chr24_+_39034090 | 1.52 |
ENSDART00000185763
|
capn15
|
calpain 15 |
| chr16_-_14353567 | 1.50 |
ENSDART00000139859
|
itga10
|
integrin, alpha 10 |
| chr4_-_685412 | 1.50 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr14_-_31814149 | 1.48 |
ENSDART00000173393
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr22_-_3344613 | 1.47 |
ENSDART00000165600
|
tbxa2r
|
thromboxane A2 receptor |
| chr25_+_15841670 | 1.47 |
ENSDART00000049992
|
syt9b
|
synaptotagmin IXb |
| chr15_+_33989181 | 1.47 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr9_-_18814737 | 1.47 |
ENSDART00000131267
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr8_-_20138054 | 1.47 |
ENSDART00000133141
ENSDART00000147634 ENSDART00000029939 |
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr5_+_1515938 | 1.46 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr15_-_43995028 | 1.46 |
ENSDART00000172485
ENSDART00000186320 |
nlrc3l
|
NLR family, CARD domain containing 3-like |
| chr3_+_22935183 | 1.45 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of myf6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.4 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.9 | 2.7 | GO:0035992 | tendon formation(GO:0035992) |
| 0.9 | 2.6 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.8 | 6.8 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.8 | 3.1 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.6 | 3.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.6 | 1.9 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.6 | 1.8 | GO:0009838 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.6 | 4.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.6 | 1.7 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.6 | 2.2 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.6 | 4.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.5 | 3.7 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.5 | 1.5 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.5 | 1.4 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.5 | 0.5 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.5 | 1.4 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.5 | 1.4 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.4 | 1.3 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.4 | 1.3 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.4 | 3.4 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.4 | 1.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 2.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.4 | 1.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.4 | 1.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.4 | 2.6 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.4 | 1.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.4 | 1.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 4.0 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.4 | 1.4 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.4 | 1.1 | GO:0036135 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.3 | 1.7 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.3 | 2.7 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.3 | 2.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.3 | 3.8 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.3 | 1.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 1.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.3 | 0.9 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.3 | 0.3 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.3 | 1.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 0.9 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.3 | 1.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 3.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.3 | 1.4 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.3 | 0.9 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.3 | 2.5 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 2.2 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.3 | 0.8 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.3 | 1.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.3 | 3.0 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 2.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 0.8 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 1.5 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.0 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 7.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.2 | 0.9 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 4.0 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 6.6 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.2 | 0.9 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.2 | 0.6 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 3.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.2 | 1.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.2 | 1.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 2.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 1.1 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.2 | 0.9 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 0.9 | GO:0014005 | microglia development(GO:0014005) |
| 0.2 | 1.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 0.6 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 | 1.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.2 | 2.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 2.4 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 2.9 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 | 0.7 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.2 | 2.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 1.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.2 | 0.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 0.6 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.2 | 2.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 0.8 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 1.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 1.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 9.5 | GO:0050922 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.7 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.1 | 0.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.4 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 1.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 2.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.5 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.0 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 1.1 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 2.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 3.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.5 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.1 | 0.6 | GO:0031650 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.7 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 1.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 1.5 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.8 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 1.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 5.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 1.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 3.1 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.1 | 2.9 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.8 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.7 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 3.6 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 9.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 2.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.0 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.4 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.1 | 1.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.5 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.5 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 1.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.8 | GO:0043507 | activation of JUN kinase activity(GO:0007257) regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.4 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:0071698 | olfactory placode development(GO:0071698) |
| 0.1 | 1.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.3 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.8 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.6 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 1.5 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.1 | 0.4 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 2.7 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 0.8 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 4.0 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.4 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.2 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.8 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.6 | GO:0035909 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.7 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.5 | GO:0000730 | DNA recombinase assembly(GO:0000730) |
| 0.1 | 0.5 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 1.2 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 1.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 4.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 6.9 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.1 | 0.3 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.1 | 0.7 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.5 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.2 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.1 | 1.2 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.1 | 0.7 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.2 | GO:0048557 | embryonic digestive tract morphogenesis(GO:0048557) |
| 0.1 | 0.6 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 5.2 | GO:0016571 | histone methylation(GO:0016571) |
| 0.1 | 0.6 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.2 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 4.8 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 0.9 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.8 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 1.1 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.3 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 1.7 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 0.3 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.9 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.1 | 1.2 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 2.0 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 2.3 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.3 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0090317 | negative regulation of protein transport(GO:0051224) negative regulation of intracellular protein transport(GO:0090317) negative regulation of establishment of protein localization(GO:1904950) |
| 0.0 | 1.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.0 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 2.0 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.3 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.4 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 10.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 2.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 2.1 | GO:0042176 | regulation of protein catabolic process(GO:0042176) |
| 0.0 | 1.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 1.6 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.2 | GO:0006266 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 5.5 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 1.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.5 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 1.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 3.0 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 1.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 3.0 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.4 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.8 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 1.5 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 2.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.3 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 2.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.6 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 1.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.4 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 1.3 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 1.2 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 2.1 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0044805 | macromitophagy(GO:0000423) late nucleophagy(GO:0044805) |
| 0.0 | 1.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 1.5 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.9 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.1 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.5 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.0 | 0.1 | GO:0098920 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 1.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0045669 | positive regulation of osteoblast proliferation(GO:0033690) positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.7 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.1 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0051236 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 1.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.3 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.5 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.5 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.0 | 0.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.7 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.5 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 2.0 | GO:0002757 | immune response-activating signal transduction(GO:0002757) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 1.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 2.8 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.4 | GO:0031673 | H zone(GO:0031673) |
| 0.8 | 2.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.8 | 3.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.6 | 11.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.5 | 2.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.4 | 2.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.4 | 1.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 1.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 2.0 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.4 | 2.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.4 | 1.9 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.4 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.4 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 2.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 13.6 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.3 | 2.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 0.9 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.3 | 2.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.3 | 3.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 1.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 1.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 2.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 4.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 3.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 4.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 0.7 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 0.9 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.2 | 0.9 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.0 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 2.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 1.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 3.9 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 1.4 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.0 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 2.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 0.5 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.2 | 1.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 2.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 3.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.5 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 1.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 8.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 1.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.2 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 2.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.9 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 12.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 0.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 2.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 5.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.8 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 2.0 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 6.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 5.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 4.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 2.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.0 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 2.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 3.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 2.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 1.7 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 3.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 7.9 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.1 | GO:0044304 | main axon(GO:0044304) |
| 0.0 | 1.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 5.5 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 2.3 | GO:0099513 | supramolecular fiber(GO:0099512) polymeric cytoskeletal fiber(GO:0099513) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 1.1 | 18.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.9 | 2.7 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.8 | 4.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.7 | 4.1 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.5 | 7.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.5 | 4.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.4 | 1.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.4 | 1.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 1.4 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.3 | 1.0 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.3 | 1.0 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.3 | 1.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 4.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.5 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.3 | 6.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.3 | 2.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 2.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.3 | 1.4 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.3 | 1.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 3.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 7.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 2.6 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.3 | 1.6 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 3.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 2.0 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.2 | 1.5 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.2 | 1.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 4.0 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 0.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 1.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 3.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 3.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 1.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 2.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.6 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 0.5 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.2 | 4.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 0.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 1.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 2.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 2.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.7 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 1.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.5 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 2.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 2.7 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 2.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.1 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 1.8 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 2.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.0 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 2.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 1.0 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 1.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 1.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.8 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 2.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 6.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 1.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 1.2 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.1 | 4.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 5.3 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.9 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.5 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.1 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 2.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.0 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 12.6 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 1.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 1.1 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.4 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 10.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 9.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.8 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 17.2 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:1901338 | catecholamine binding(GO:1901338) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.5 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 3.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 2.1 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 4.3 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 4.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 1.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 1.4 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 5.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 3.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 3.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 4.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 8.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 2.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.2 | 3.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 2.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 8.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.1 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 5.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.1 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.0 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 4.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 4.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.4 | 7.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 3.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 1.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 4.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 2.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 2.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 4.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.2 | 2.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 7.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 3.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.7 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.1 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 2.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.7 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 1.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 3.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 1.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.9 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 2.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.3 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.5 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 1.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 2.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |